Engineering bacterium and application thereof in preparation of atorvastatin drug intermediate
An engineering bacteria and purpose technology, applied in the field of biopharmaceuticals, can solve the problem of low activity of the substrate halohydrin, and achieve the effects of simple operation, mild reaction conditions, and improved conversion rate and purity.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
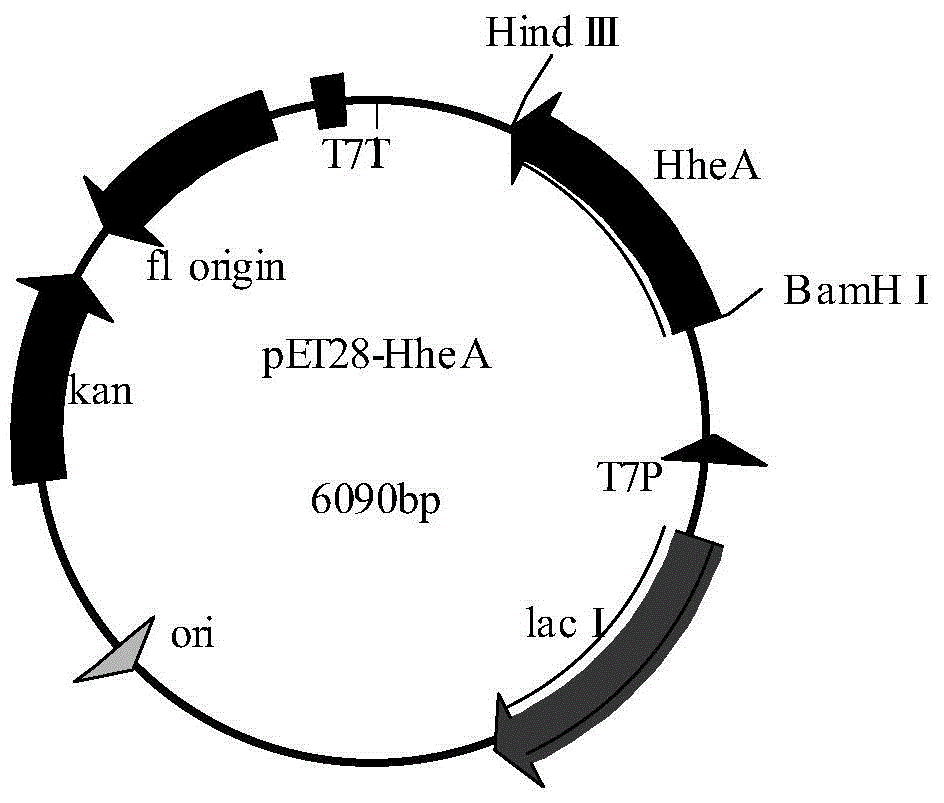
[0048] Construction of embodiment 1 plasmid pET28-HheA
[0049] The halohydrin dehalogenase gene is the hhea gene, which is a fully synthetic gene and connected to the pMD18-HHEA plasmid. (The gene is derived from Arthrobacter sp.AD2, and the Genebank accession number is AF397297)
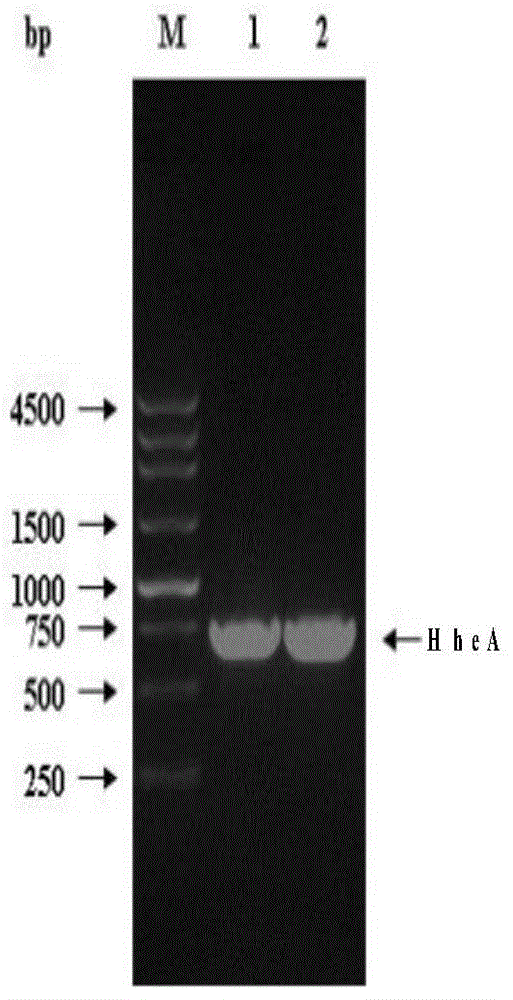
[0050] The HheA gene was cloned with primers F_HheA / R_HheA to obtain a 735bp HheA gene (SEQ ID No.3). Nucleic acid electrophoresis to verify gene size, such as figure 1 .
[0051] The sequence of primer F_HheA is:
[0052] 5'-CGCGGATCCATGGTCATCGCACTGGTGACGCA (SEQ ID No. 1);
[0053] The sequence of primer R_HheA is:
[0054] 5'-CCCAAGCTTTTACGGCAGATAGCCGCCGGTAAA (SEQ ID No. 2).
[0055] BamHI and HindIII double-enzyme digested HheA gene, recovered the gene band after digestion, BamHI and HindIII double-enzyme digested pET-28a(+) plasmid, recovered the plasmid band after digestion, and digested HheA gene and enzyme The excised pET-28a(+) plasmid was ligated with ligase and transformed into the...
Embodiment 2
[0056] Construction and induced expression of embodiment 2 genetically engineered bacteria
[0057] The plasmid pET28-HheA constructed in Example 1 was used to transform the expression host Escherichia coli BL21(DE3). Use primers F_HheA / R_HheA to do colony PCR to verify the transformed recombinants. The verified genetically engineered bacteria is Eco HheA. Inoculate Eco HheA into 3-5mL liquid LB test tube medium containing kanamycin resistance, activate it on a shaking table at 37°C for 12 hours, and transfer the culture obtained after activation to 1% transfer amount containing In the kanamycin-resistant liquid LB shake flask medium, the fermentation medium was shaken and cultured at a constant temperature for 3 hours, and the culture conditions were 37° C. and 200 rpm. When the cell concentration grows to OD 600 When =0.8, add 0.5mM IPTG (final concentration), induce at 18°C for 13h, centrifuge at 10,000g for 5min to collect the cells, wash once with pH=7.0 HEPES buffer...
Embodiment 3
[0058] Construction and induced expression of embodiment 3 genetically engineered bacteria
[0059] The plasmid pET28-HheA constructed in Example 1 was used to transform the expression host Escherichia coli BL21(DE3). Use primers F_HheA / R_HheA to do colony PCR to verify the transformed recombinants. The verified genetically engineered bacteria is Eco HheA. Inoculate Eco HheA into 3-5mL liquid LB test tube medium containing kanamycin resistance, activate it on a shaking table at 35°C for 12 hours, and transfer the culture obtained after activation to 1% transfer amount containing In the kanamycin-resistant liquid LB shake flask medium, the fermentation medium was shaken at constant temperature for 3 hours, and the culture conditions were 35° C. and 200 rpm. When the cell concentration grows to OD 600 When = 1.2, add 0.1mM IPTG (final concentration), induce at 22°C for 16h, centrifuge at 10,000g for 5min to collect the cells, wash once with pH=9.0 HEPES buffer, discard the su...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com