A bac-fish method for simultaneous identification of complete sets of chromosomes in each subgroup in the background of ad or ade in cotton
A chromosome and background technology, applied in the field of molecular cytogenetics, can solve the problems of workload, technical requirements, reagent cost, etc.
Inactive Publication Date: 2017-05-24
INST OF COTTON RES CHINESE ACAD OF AGRI SCI
View PDF0 Cites 0 Cited by
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
View moreImage
Smart Image Click on the blue labels to locate them in the text.
Smart ImageViewing Examples
Examples
Experimental program
Comparison scheme
Effect test
Embodiment 1
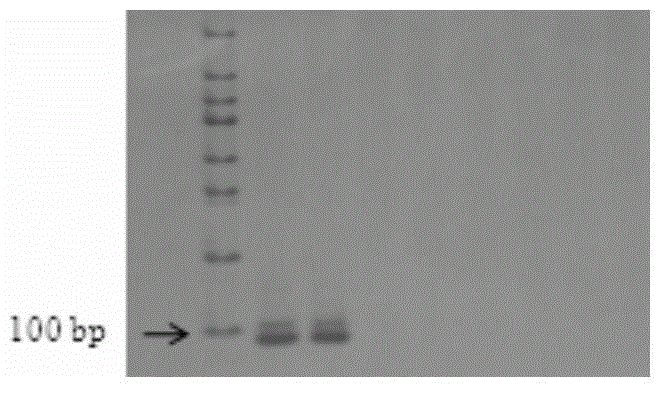
[0022] Example 1 BAC clone 57I23 was used as a probe to hybridize with two diploids, five tetraploids, and one hexaploid hybrid somatic metaphase chromosome of cotton.
[0023] 1 Materials and methods
[0024] 1.1 Experimental materials
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More PUM
 Login to View More
Login to View More Abstract
The invention belongs to the field of molecular cytogenetics and particularly relates to a BAC-FISH method for simultaneously identifying whole sets of chromosomes of subgroups under a gossypium AD / ADE background. The method comprises the following steps: carrying out BAC-FISH on different cotton seeds by using BAC cloned 57I23 originated from an Africanum BAC library, and simultaneously identifying chromosomes of an AD genome and AD sub genome of cotton obviously; by using BAC-DNA as a probe, covering most regions out of ends of all chromosomes of the subgroup A in five gossypium tetraploids as well as a small region near the middle part of the all chromosomes of the subgroup D by a hybrid signal; and in allohexaploid hybrids AADDEE hybridized by upland cotton and G.stocksii (E1) in the group E, as the hybrid signal is similar to the tetraploid, obviously differentiating the hybrid signal from the subgroup E (without a signal) so as to simultaneously identify three sets of chromosomes of subgroups (A,D,E). By adopting the BAC-FISH method through BAC cloning, the whole sets of chromosomes of subgroups under the gossypium AD / ADE background can be simultaneously identified.
Description
technical field [0001] The invention belongs to the field of molecular cytogenetics, and specifically relates to a BAC-FISH method for simultaneously identifying a complete set of chromosomes of each subgroup under the background of AD or ADE of cotton. Background technique [0002] Fluorescence in situ hybridization (FISH) technology can locate DNA sequence on chromatin, chromosome or DNA fiber, and is an effective and precise molecular cytogenetics method. Plant genome sequencing is required for functional genome research, and the divergence between genetic distance and physical distance limits the application of linkage maps in guiding genome sequence assembly and map-based cloning. The cytogenetics map represents the actual position of the DNA sequence on the chromosome, and is one of the types of genome physical maps, so it is also the basis for genome sequencing and sequence assembly. Direct positioning of DNA clones on chromosomes by BAC-FISH is currently one of the ...
Claims
the structure of the environmentally friendly knitted fabric provided by the present invention; figure 2 Flow chart of the yarn wrapping machine for environmentally friendly knitted fabrics and storage devices; image 3 Is the parameter map of the yarn covering machine
Login to View More Application Information
Patent Timeline
 Login to View More
Login to View More Patent Type & Authority Patents(China)
IPC IPC(8): C12Q1/68
CPCC12Q1/6841C12Q1/6895C12Q2561/101
Inventor 刘玉玲赵艳艳刘方周忠丽蔡小彦王星星王春英王玉红彭仁海王坤波
Owner INST OF COTTON RES CHINESE ACAD OF AGRI SCI
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com