Bacillus licheniformis expression host
A technology of Bacillus licheniformis and host, applied in the direction of bacteria, enzymes, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
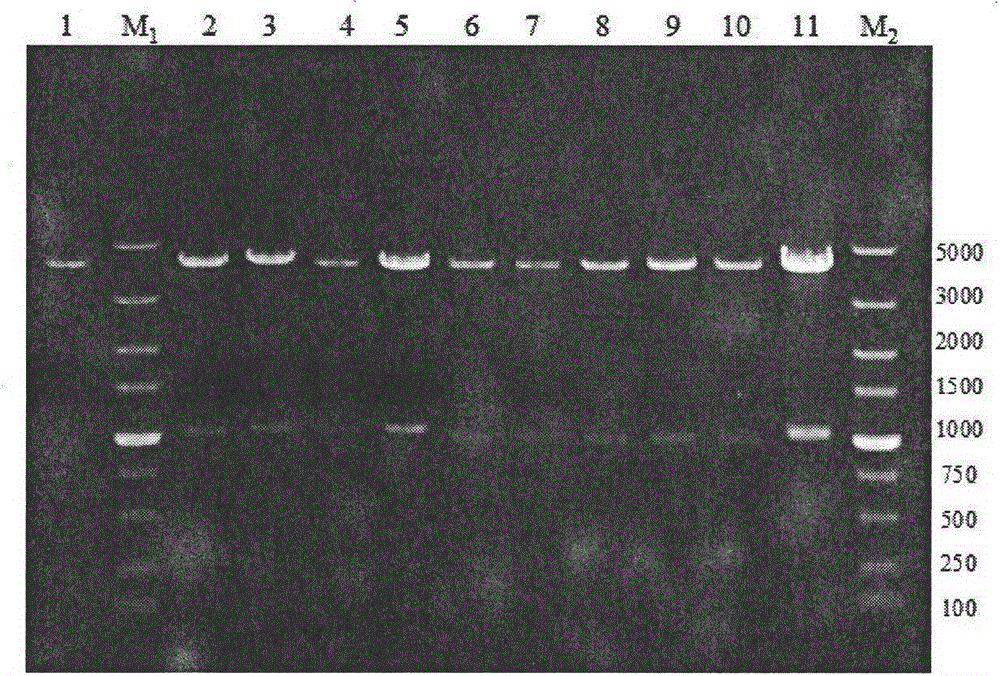
[0054] Construction of embodiment 1 protease deletion strain BL10
[0055] 1. Annotation of Bacillus licheniformis WX-02 protease (or peptidase)
[0056] Using all the known and putative proteases or peptidases of Bacillus licheniformis listed in the MEROPS database, Blast found the corresponding protease or peptidase genes in the WX-02 genome sequence, and then predicted them with Signal-3L and Cell-PLoc Its location in the cell, extracellular (including cell wall), intracellular or cell membrane. Then, using the reported extracellular proteomics results of B. licheniformis as a reference, some proteases or peptidases that can be secreted extracellularly without signal peptides were corrected, and their protein molecular weights were calculated using EditSeq. Through the analysis of the B. licheniformis genome sequence obtained by sequencing, it is concluded that there are 166 known and putative proteases (or peptidases) in the genome, among which the (known and putative) pr...
Embodiment 2
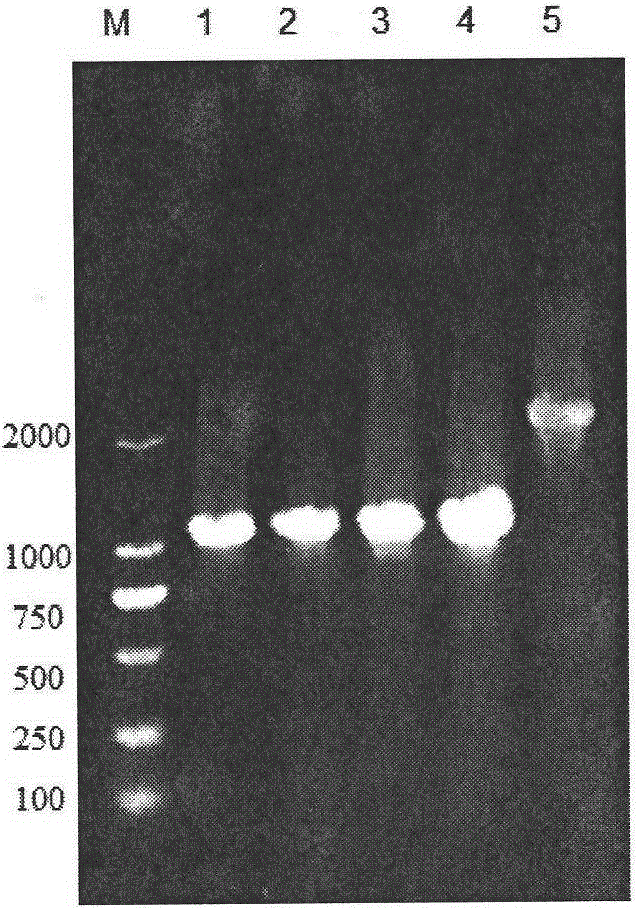
[0075] High-efficiency secretory expression of embodiment 2α-amylase
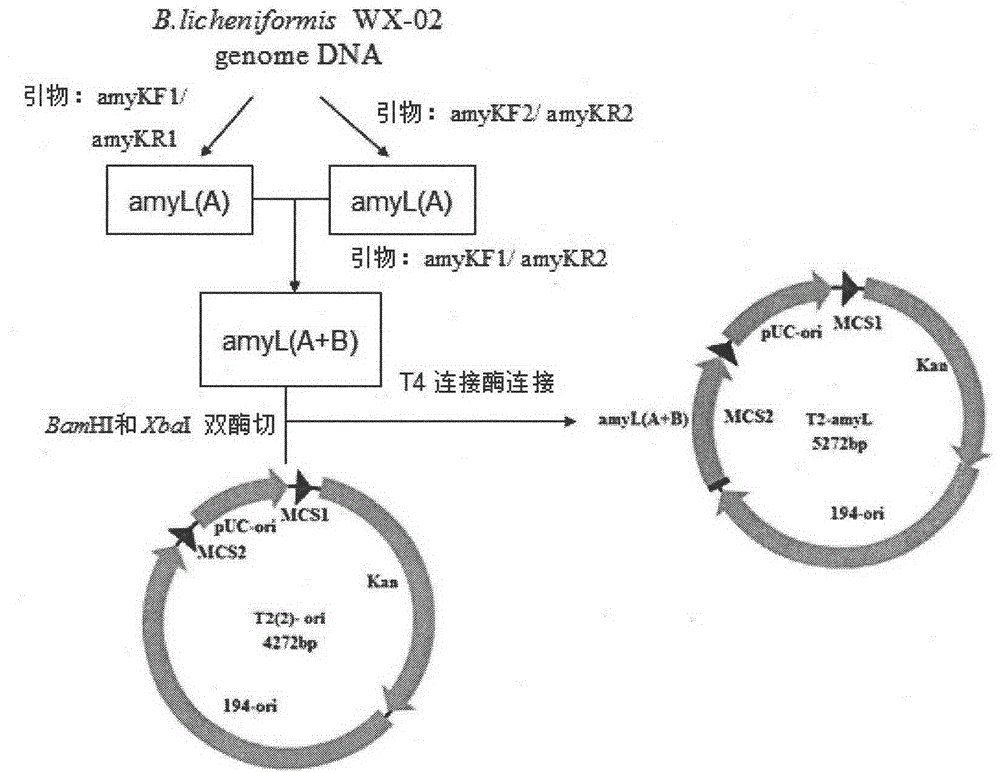
[0076] The construction process of B. licheniformis α-amylase expression plasmid pP43SAT is as follows: Figure 8 shown. A pair of primers P43F and P43R1 were designed according to the sequence of the P43 promoter on the B.subtillis168 genome published by NCBI,
[0077] P43F: 5'-GGAATTCTGATAGGTGGTATGTTTTCG-3' (EcoRI)
[0078] P43R1: 5'-AAGCCGTTTTTGTTGTTTCATTTCATGTGTACATTCCTCTC-3'
[0079] The promoter P43 fragment was amplified by PCR with a size of 305bp.
[0080] According to the amyL gene sequence (see SEQ ID NO.9) in the whole genome sequence of B. licheniformis WX-02 measured in this experiment, the accession number on NCBI is MUY_00877, and a pair of primers PamyLF1 and PamyTR were designed,
[0081] PamyLF1:
[0082] 5'-GAGAGGAATGTACACATGAAATGAAACAACAAAAACGGCTT-3'
[0083] PamyTR: 5'-GAAGATCT CGCAATAATGCCGTCGCACTG-3'(BglII)
[0084] Using the genomic DNA of B. licheniformis WX-02 as a template...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com