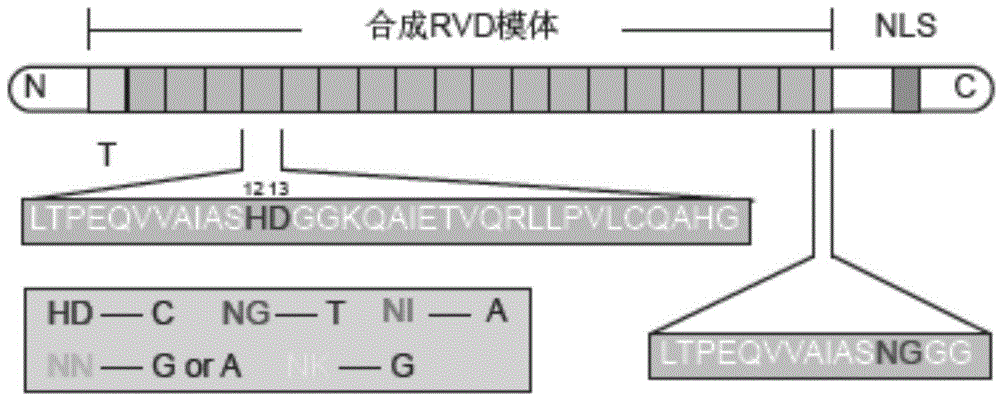
Modular construction of synthetic gene route in mammalian cell by TALE transcription suppressor
A cell, coding gene technology, applied in the field of modular construction of synthetic gene circuits, can solve problems such as low inhibition efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0126] The method of transfecting cells with plasmids in Example 1 and Example 2: take a 24-well plate and inoculate 0.5 mL of HEK293 cell suspension (containing 6×10 4 HEK293 cells), cultured for 24 hours, replaced with new DMEM medium, and then carried out plasmid transfection.
[0127] Example 1, Functional verification and specificity analysis of TALER protein
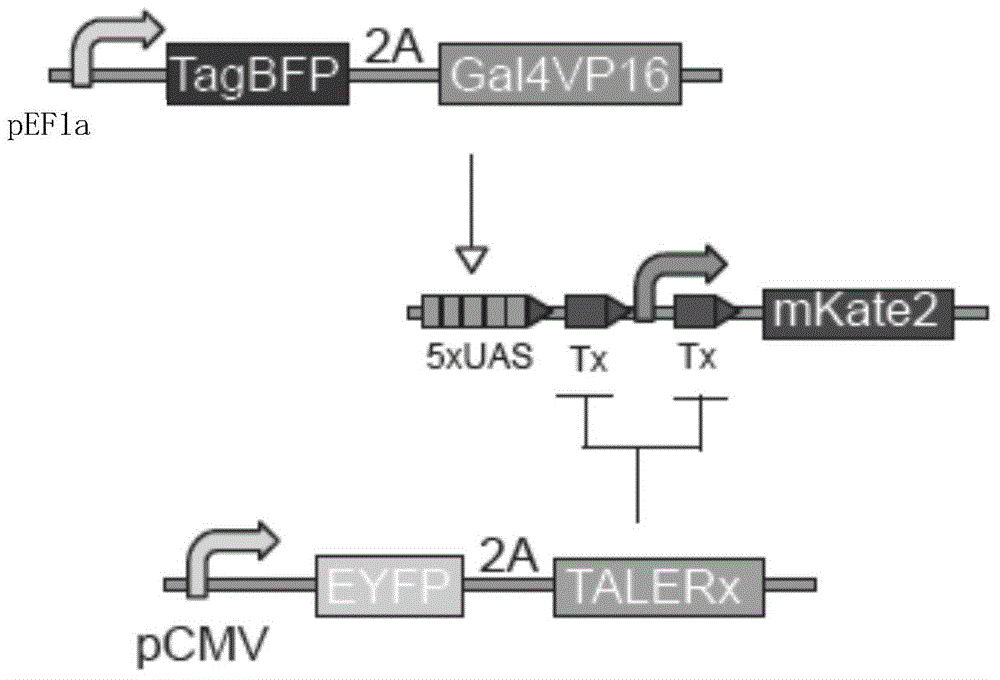
[0128] The schematic diagram of the action mechanism of pCMV-TALERx plasmid, pTx+Tx+ plasmid and pEF1a-TagBFP-2A plasmid is shown in figure 2 . Under the action of the pEF1a promoter, TagBFP and Gal4 / vp16 are expressed (the 2A connecting peptide between TagBFP and Gal4 / vp16 is a self-splicing peptide, so TagBFP can represent the expression level of Gal4 / vp16). Gal4 / vp16 activated the 5×UAS sequence, thereby activating the transcriptional initiation of the CMVmini promoter, and mKate2 was expressed. Under the action of the CMV promoter, EYFP and TALER1 proteins are expressed (the 2A connecting peptide between EY...
Embodiment 2
[0195] Embodiment 2, further ductility research
[0196] The pEF1a-TagBFP-2A plasmid is the pEF1a-TagBFP-2A plasmid in Example 1.
[0197] The pCMV-TALER1 plasmid is the pCMV-TALER1 plasmid in Example 1.
[0198] The pCMV-TALER2 plasmid is the pCMV-TALER2 plasmid in Example 1.
[0199] The pCMV-TALER4 plasmid is the pCMV-TALER4 plasmid in Example 1.
[0200] The pCMV-TALER5 plasmid is the pCMV-TALER5 plasmid in Example 1.
[0201] The pCMV-TALER32 plasmid is the pCMV-TALER32 plasmid in Example 1.
[0202] The pT1+T1+72-DsRed plasmid is shown in SEQ ID NO:54. In sequence 54, nucleotides 2441-2533 from the 5' end are 5×UAS sequences, nucleotides 2549-2562 are T1 sequences (target sequence of TALER1 protein), nucleotides 2569-2628 It is a CMVmini promoter, the 2635-2648th nucleotide is the T1 sequence, and the 2668-3345th nucleotide is the coding gene of DsRed (red fluorescent protein).
[0203] The pT1+T2+72-DsRed plasmid is shown in SEQ ID NO:55. In sequence 55, nucleoti...
Embodiment 3
[0245] Example 3, Modular construction of the gene circuit of the TALER protein cascade
[0246] The pCAG-rtTA-2A-Gal4 / vp16 plasmid is shown in SEQ ID NO:73. In sequence 73, nucleotides 4253-4930 from the 5' end are the CAG promoter, nucleotides 6004-6747 are the gene encoding rtTA, and nucleotides 6748-6813 are the gene encoding the 2A linker peptide , the 6820-7503 nucleotides are the coding gene of Gal4 / vp16.
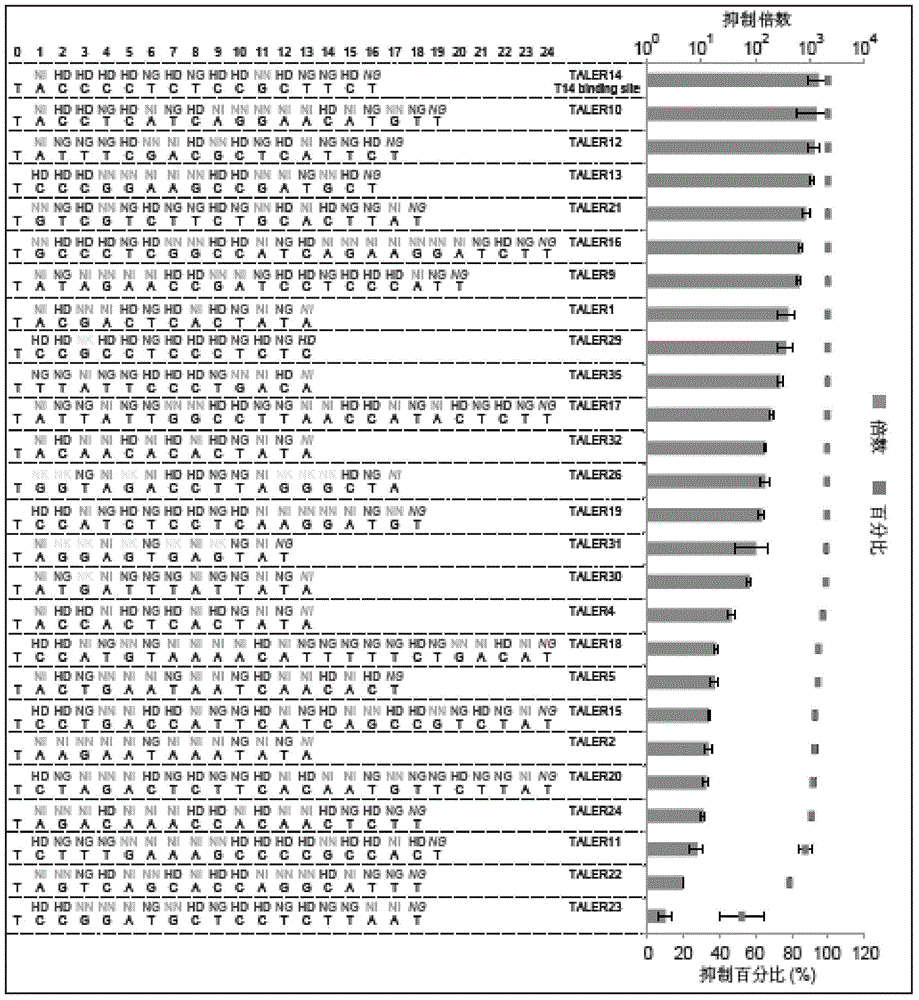
[0247] The pT14+T14+72-mKate2 plasmid is shown in SEQ ID NO:74. In sequence 74, nucleotides 4275-4367 from the 5' end are 5×UAS sequences, nucleotides 4383-4399 are T14 sequences (target sequence of TALER14 protein), nucleotides 4406-4465 It is a CMVmini promoter, the 4472-4488th nucleotide is the T14 sequence, and the 4538-5243rd nucleotide is the coding gene of mKate2. The pT14+T14+72-mKate2 plasmid has two TALER14 protein binding sites (pT14BS2).
[0248] The pT14+T14x3+72-mKate2 plasmid is shown in SEQ ID NO:75. In sequence 75, nucleotides 69-161 from the 5'...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com