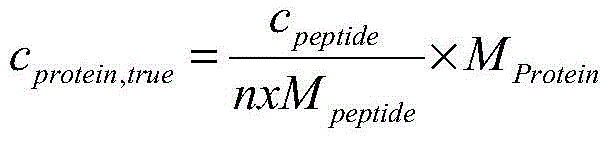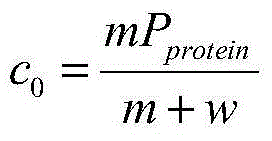Method for accurately testing digestion efficiency of proteins in matrix
A protein enzyme, accurate determination technology, applied in measuring devices, material analysis by electromagnetic means, instruments, etc., can solve the problems of enzyme cleavage efficiency difficult to reach 100%, missed peptide fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0124] A method for accurately determining the enzyme cleavage efficiency of a protein in a substrate, characterized in that it comprises the following steps:
[0125] (1) Screen the specific peptides of the target protein, use protease to theoretically digest the target protein, and obtain each theoretically digested peptide; use the Blast tool to search the obtained theoretically digested peptides one by one, and determine the target protein specific enzyme-cut peptides;
[0126] (2) Specific peptides are synthesized from amino acids by chemical synthesis; isotope-labeled specific peptides are synthesized from isotope-labeled amino acids, and the synthesized peptides are purified by reverse-phase high-performance liquid chromatography to obtain pure products , using the isotope dilution mass spectrometry method based on amino acid analysis to determine the mass fraction P of the target peptide in the synthetic peptide peptide ;
[0127] (3) Obtain the pure product of the t...
Embodiment 2
[0176] 1. Instruments used:
[0177] Analytical balance: (METTLER TOLEDO XS205 type, the maximum weighing capacity is 81g, the precision is 0.01mg, the United States); (METTLER TOLEDO XP56 type, the maximum weighing capacity is 52g, the precision is 0.001mg, the United States);
[0178] Constant temperature shaker (S16R-2 type, SHEL LAB, USA);
[0179] Oven (UFE500 type, MEMMERT, the United States);
[0180] Liquid chromatography-mass spectrometry (TSQ Vantage Triple Quard LC / MS, Thermo, USA);
[0181] High-speed centrifuge (type 3K15, Sigma, Germany);
[0182] Ultrapure water system (Milli-Q type, MILLIPORE, USA);
[0183] Centrifugal ultrafiltration tubes (Amicon ultra-4 type, Millipore, USA).
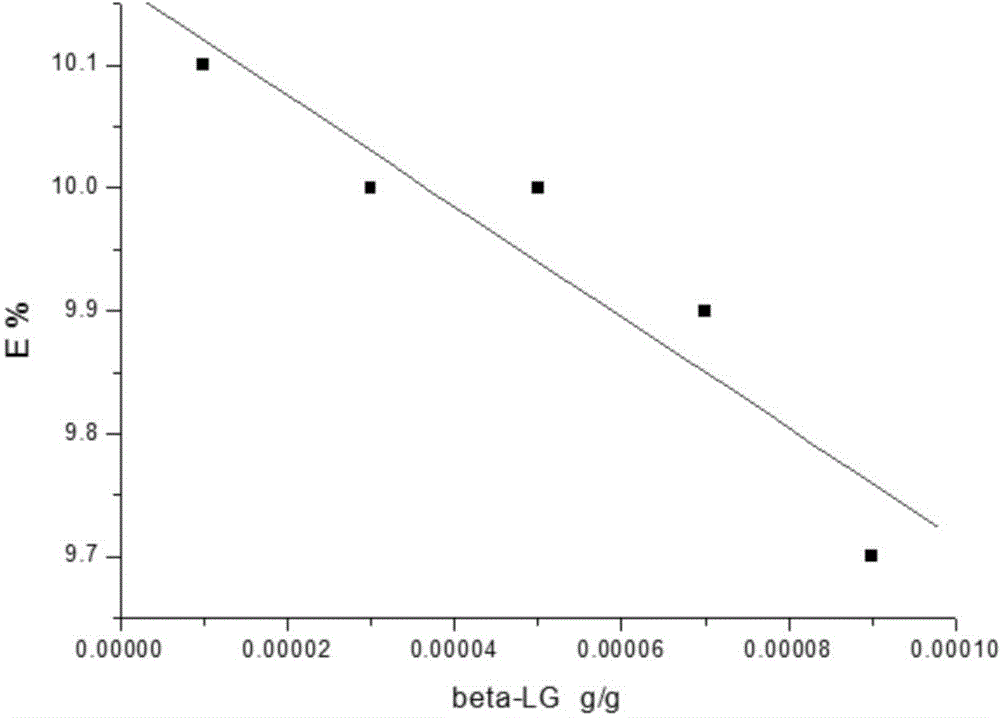
[0184] 2. Accurate determination of β-lactoglobulin digestion efficiency in milk powder, including the following steps:
[0185] (1) Search the database by means of bioinformatics, and select TPEVDDEALEK (TK-11) as the specific peptide for β-lactoglobulin isotope dilution mass spe...
PUM
| Property | Measurement | Unit |
|---|---|---|
| quality score | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com