Environment DNA identification method for fish community structure researching
A technology of community structure and identification method, applied in the field of molecular ecology, can solve the problems of high fishing intensity requirements, difficult to guarantee accuracy, difficult to achieve good results, etc., and achieve good versatility and applicability, sensitivity and accuracy. Improve and save human and financial resources
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] Screening of universal primers for the most ideal fish environment:
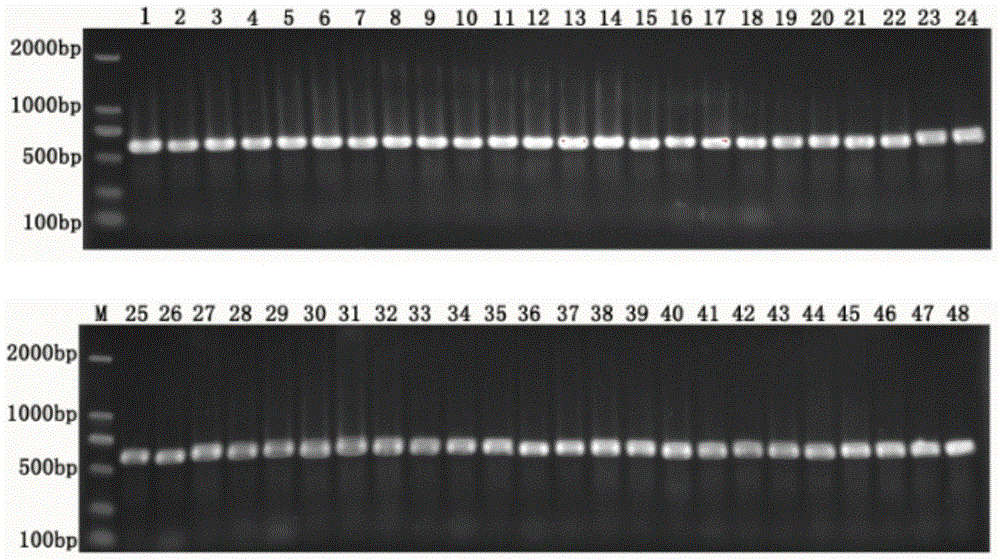
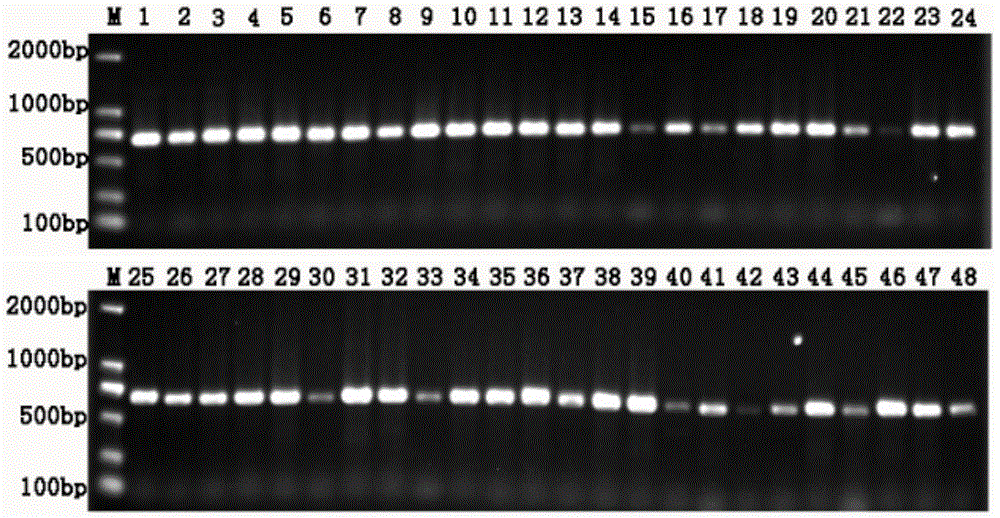
[0034]Four genes of fish mitochondria were selected: 16S rDNA (SEQ ID NO:1; SEQ ID NO:2), COI (SEQ ID NO:3; SEQ ID NO:4), Cytb1 (SEQ ID NO:5; SEQ ID NO: 6), Cytb4 (SEQ ID NO: 7; SEQ ID NO: 8), D-Loop (SEQ ID NO: 9; SEQ ID NO: 10) 5 universal primers of partial sequences, which are shown in Table 1 showed that the fish samples and environmental water samples from Qiandao Lake in Hangzhou, Zhejiang Province were selected as the research samples for the comparative verification experiment, and the generality of primers for 48 main fish species in Qiandao Lake was compared. After the amplification of 48 kinds of fish, only 16s The rDNA amplification effect is the best, 48 and all amplified bright bands, such as Figure 2 ~ Figure 6 As shown; when the 10 environmental DNA samples of Qiandao Lake were amplified, only the 16s rDNA had the best amplification effect, and a bright band was obtained, which can ...
Embodiment 2
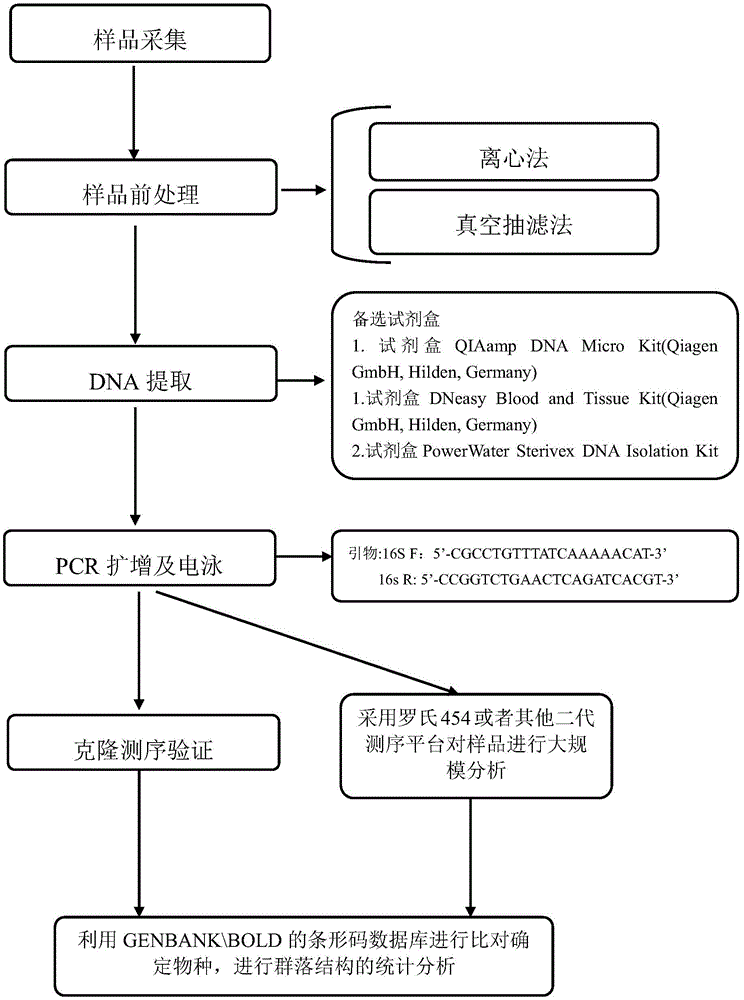
[0039] The experimental process is as follows figure 1 As shown, at the same time, two points were randomly selected in the upper reaches of Qiandao Lake to collect water samples. Each sample was 1 L, put in an incubator with ice, and the water samples were filtered within 24 hours. The filter membrane pore size was 3 μm. The environmental DNA in the filter membrane was extracted using the kit QIAamp DNA Micro Kit produced by QIAGEN. The extracted DNA was dissolved in TE buffer and stored at -20°C.
[0040] The 16s rDNA sequence of SEQ ID NO: 1 was used as a universal primer for amplification, and the PCR products were detected by gel electrophoresis. Each PCR product was taken in 1% agarose gel, and the voltage was 100V. Electrophoresis was performed for 60 minutes. Ethidium staining was performed for 8 minutes, and finally photographed on a gel imaging system, and the amplification effect of each pair of primers was counted.
[0041] The environmental samples amplified by ...
Embodiment 3
[0045] After selecting 5 environmental samples from Qiandao Lake for environmental DNA extraction and 16s rDNA amplification, 5 barcodes were designed at both ends of the primer sequences (see Table 3), and the environmental samples were amplified using primers 16sF and 16sR with added Barcode sequences. The Roche 454 next-generation sequencer sequenced the samples, and the statistics of the results are shown in Table 4: 13875-28022 sequences were obtained from the five samples, and the average sequence length was about 597.28bp-598.55bp. All of them were 16s rDNA sequences of Qiandao Lake fishes after comparison.
[0046] Table 3 Barcode design table of 5 samples
[0047] Sample
A1
ACACGCTG
A3
ACATGTCA
A5
ACGAGTGC
A7
ACGCGATA
A9
ACTCGCAC
[0048] Table 4 Statistics of sequencing results
[0049] Sample
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com