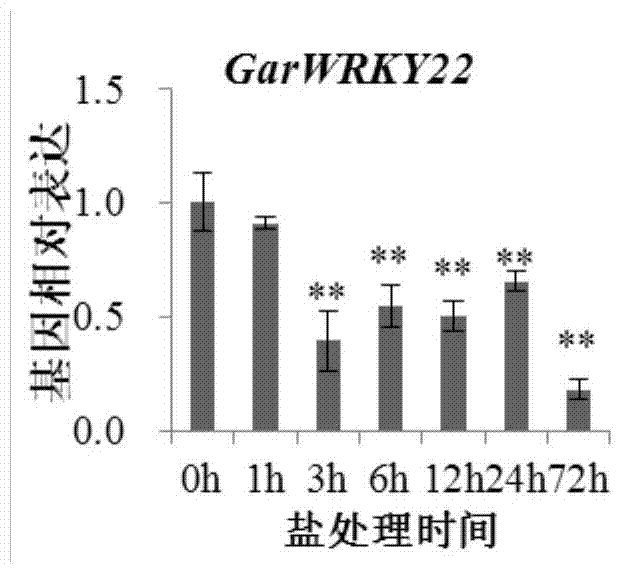
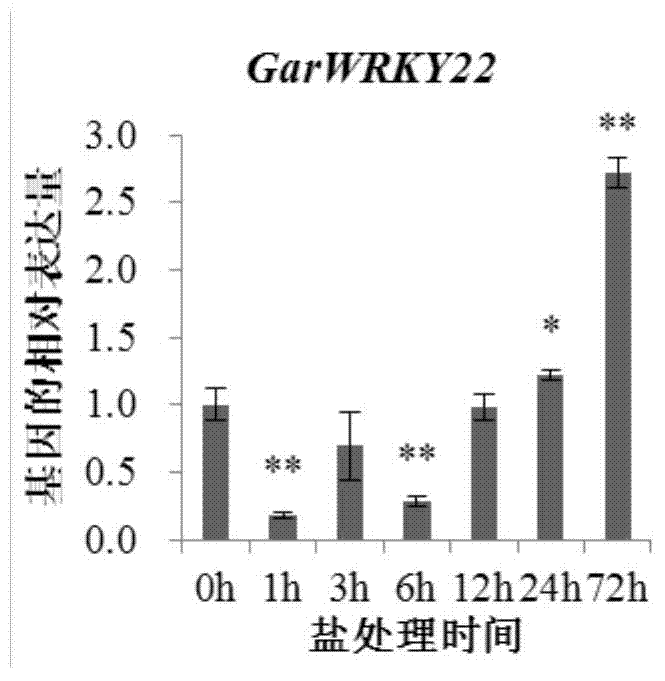
Cotton WRKY transcription factor GarWRKY22 for regulating salt tolerance of plants and application thereof
A technology of transcription factor and salt tolerance, applied in the field of biological genetic engineering, can solve the problems of few research reports and achieve the effect of reducing salt tolerance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Embodiment 1, the acquisition of GarWRKY22 gene
[0023] 1.1 Extraction of RNA
[0024] extract RNA
[0025] (1) Take 0.5g of fresh cotton tissue, add 0.1g of cross-linked polyvinylpyrrolidone (PVPP), fully grind to powder in liquid nitrogen, quickly transfer the frozen powder into a 10ml centrifuge tube, add 5ml of CTAB extract With 500 μL of 0.1M Tris-HCl of pH 8.0, bathe in water at 65°C for 20 minutes, and mix well by inverting halfway;
[0026] (2) Add an equal volume of chloroform to mix well, and let stand in an ice bath for 10 minutes;
[0027] (3) Centrifuge at 10,000 rpm for 20 minutes at 4°C. Divide into four 1.5ml centrifuge tubes;
[0028] (4) Aspirate the supernatant, add 1 / 3 volume of 8M LiCl and mix, -70°C for 30min or -20°C overnight;
[0029] (5) Centrifuge at 10,000 rpm for 20 minutes at 4°C. Discard the supernatant, wash twice with 70% ethanol, dry the precipitate and dissolve it in 30 μL DEPC water;
[0030] (6) Add 10U DNase without RNase ac...
Embodiment 2
[0062] Embodiment 2, the construction of plant expression vector
[0063] 2.1 Construction of pCAMBIA2301-CaMV35S-GarWRKY22 plant expression vector
[0064] Plant expression vector pCAMBIA2301-CaMV35S plasmid (Feng Juan et al., 2013; Cloning and functional analysis of the protein kinase gene GarCIPK8 of the wild species of Gossypium upland). BamHI and KnpⅠ were used to digest pCAMBIA2301-CaMV35S and the target gene fragment GarWRKY22 respectively, recover the large vector fragment and the target gene fragment, and transform E. coli Trans1-T1 competent cells (purchased from Beijing Quanshijin Biology Co., Ltd.) after ligation with T4 ligase. Technology Co., Ltd.), and the plant expression vector with the gene of interest was obtained after the recombinant was identified.
[0065] Enzyme digestion of pCAMBIA2301-CaMV35S plasmid and target gene fragment
[0066] The plasmid double enzyme digestion system is as follows:
[0067]
[0068]
Embodiment 3
[0086] Embodiment 3, preparation and transformation of Agrobacterium competent
[0087] 3.1 Preparation of Competent Agrobacterium EHA105
[0088] (1) Pick a single colony of EHA105, inoculate it in 5ml LB liquid medium, and culture overnight at 28°C with shaking at 200rpm until the OD600 value is 0.4;
[0089] (2) Inoculate in 400-500ml LB medium (in a 1L Erlenmeyer flask) at a ratio of 1:100, shake the bacteria until the OD600 is 0.6-0.8, and bathe in ice for 10 minutes;
[0090] (3) Collect the bacterial solution in a pre-cooled 50ml centrifuge tube, centrifuge at 5000rpm at 4°C for 5min;
[0091] (4) Discard the supernatant, fully suspend the precipitate with sterile water, centrifuge at 5000 rpm at 4°C for 5 min; repeat this process 3 times.
[0092] (5) Add 1 ml (depending on the number of bacteria) to the washed cells to resuspend the cells containing 10% sterile glycerol.
[0093] (6) Aliquot into 50 μL tubes, quick freeze in liquid nitrogen, and store at -80°C for ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com