New oxidoreductases for enantioselective reactions
An active and functional technology, applied in the field of compositions for generating new variants-2-hydroxyacid dehydrogenase, can solve problems such as unpredictable correct alignment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
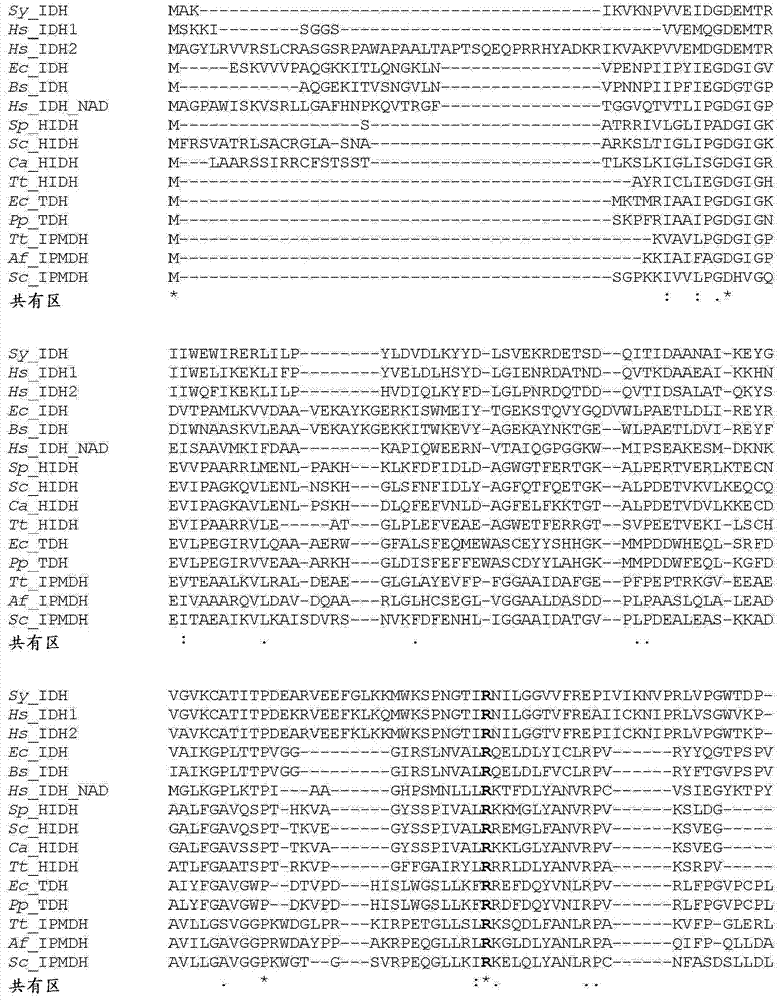
Image
Examples
Embodiment 1
[0214] Preparation of HIDH constructs
[0215] A blunt-ended 1002 base pair (bp) fragment encoding Thermophila HIDH (TTC1012) lacking the 3'-stop codon was amplified from the Thermomicrobial HB27 gDNA library (ATCC). A blunt-ended 1110 bp fragment encoding LYS12 lacking the 3'-stop codon was amplified from a S. cerevisiae gDNA library using KAPA Taq DNA polymerase (KAPA Biosystems, Woford, MA). These fragments were cloned into pTrcHis2-TOPO (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. Mutations were performed using the QuikChange Site-Directed Mutagenesis Kit (Agilent). All constructs were verified by sequencing with pTrcHis2-for and pTrcHis2-rev primers. The constructs are listed in Table 1, SEQ ID NO: 1-22.
Embodiment 2
[0217] Purification of HIDH mutants
[0218] The expression and purification procedure for homoisocitrate dehydrogenase was adapted from published methods. Lin et al., Biochemistry 46:890-898 (2007). Expression constructs were transformed into BL21-DE3 E. coli (Stratagene). Single colonies were inoculated into 5 mL LB-Amp medium starter cultures and grown with shaking (37° C. 225 rpm) for 6 h. Starter culture (5 mL) was added with 45 mL LB-Amp, shaken for 2 h, and induced with 1 mM IPTG and shaken for another 2 h. Centrifuge at 4,500×g at 4°C to collect the precipitate and dissolve in 2 mL of buffer B (500 mM NaCl, 10 mM MgCl 2 , 20 mM imidazole, 2 mM β-mercaptoethanol, 10 mM Tris, pH 7.5, supplemented with 1× EDTA-free cOmplete Mini from Roche TM protease inhibitors) and sonicated on ice for 6 cycles of 15 sec each on a Sonifer 250 (Branson). The lysate was clarified by centrifugation at 13,000 xg at 4°C. Lysates were loaded onto Ni-NTA spin columns (Qiagen) that had ...
Embodiment 3
[0220] Preparation of crude lysates of HIDH mutants
[0221] To obtain a crude lysate, 0.5 mL starter culture as described above was added to 4.5 mL LB-Amp and shaken for 2 h followed by an additional 2 h of incubation with 1 mM IPTG. The pellets from these cultures were collected by centrifugation at 4,500 xg at 4°C. These pellets were resuspended on ice in 500 μL buffer A (0.2% Triton-X 100, 1 mM PMSF, 0.5 mM EDTA, 10 mM Tris, pH 7.5) and sonicated on ice for 6 cycles of 15 s each on a Sonifer 250 (Branson) . Then, it was centrifuged down at 13,000×g 4°C for 15 minutes, and the clarified lysate was normalized to 700 ng / μL with lysis buffer and aliquoted for storage at -80°C. Protein concentrations were determined using BioRad protein assay reagents according to the manufacturer's instructions.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com