Method for accurately and rapidly identifying drug target of active compound obtained by phenotypic screening
A compound and active technology, applied in the field of medicine, can solve problems such as unknown targets, and achieve the effect of acceleration speed and precision
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
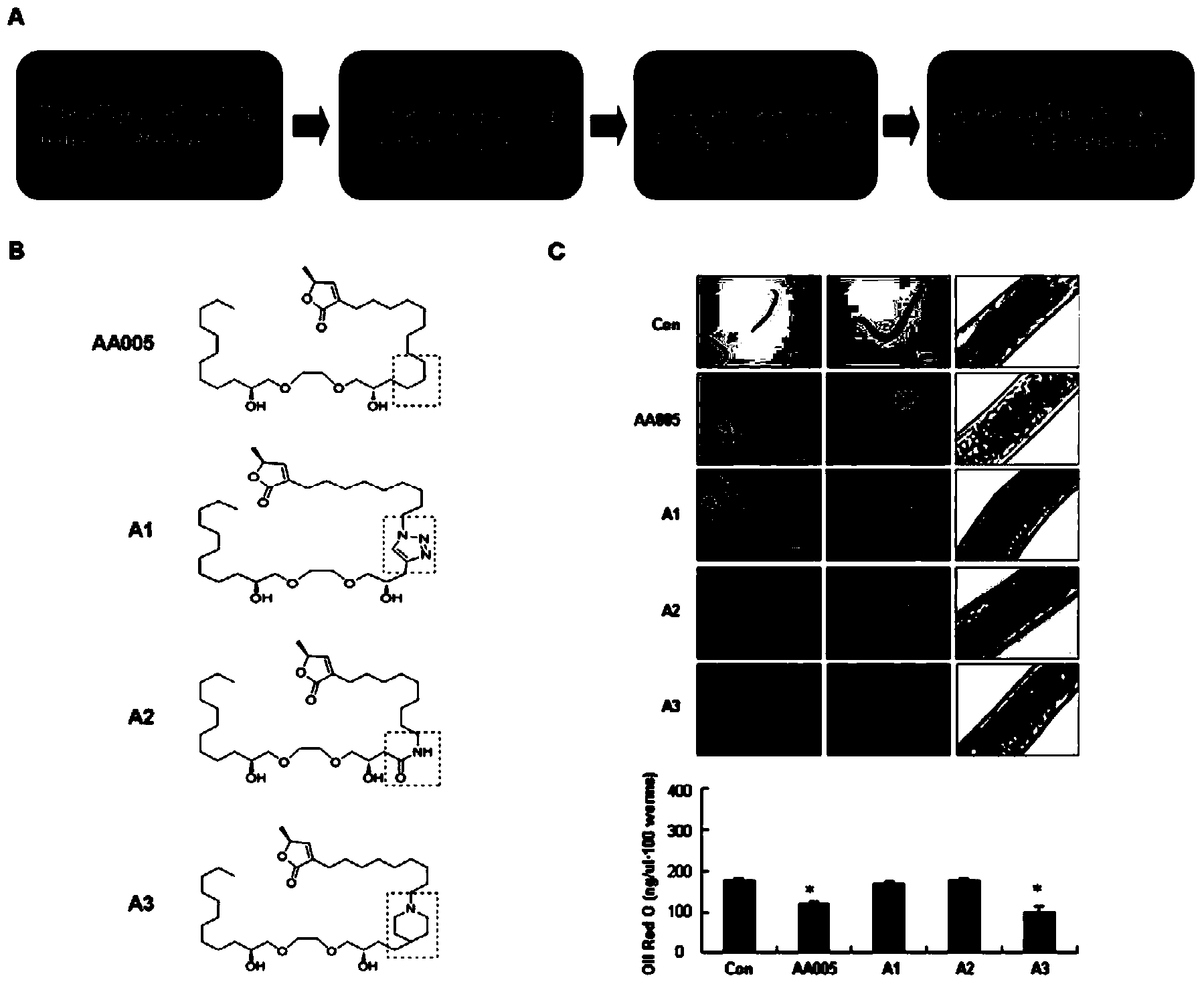
[0049] Example 1. AA005 and its structural analogs can specifically inhibit nematode body fat accumulation without toxicity
[0050] Synchronized L1-stage N2 nematodes were reared on NGM plates with OP50 bacteria for 28 hours, and the nematodes were evenly divided into six-well plates, and the nematodes were treated with 4 μM AA005, A1, A2, and A3 compounds for 14 hours, and the control group did not add drugs , and appropriate OP50 bacteria feeding. Each group of nematodes after drug treatment were placed on the NGM plate containing OP50 bacteria to recover for 16 hours, and an equal amount of nematodes were collected for staining with oil red O, and oil red O was extracted for quantification, and the effect of AA005 and its derivatives on the lipid accumulation of nematodes was observed . Observation under the microscope ( figure 1 C) Compared with the control group, the lipids in nematodes treated with compounds AA005 and A3 were significantly reduced, while the lipids of...
Embodiment 2
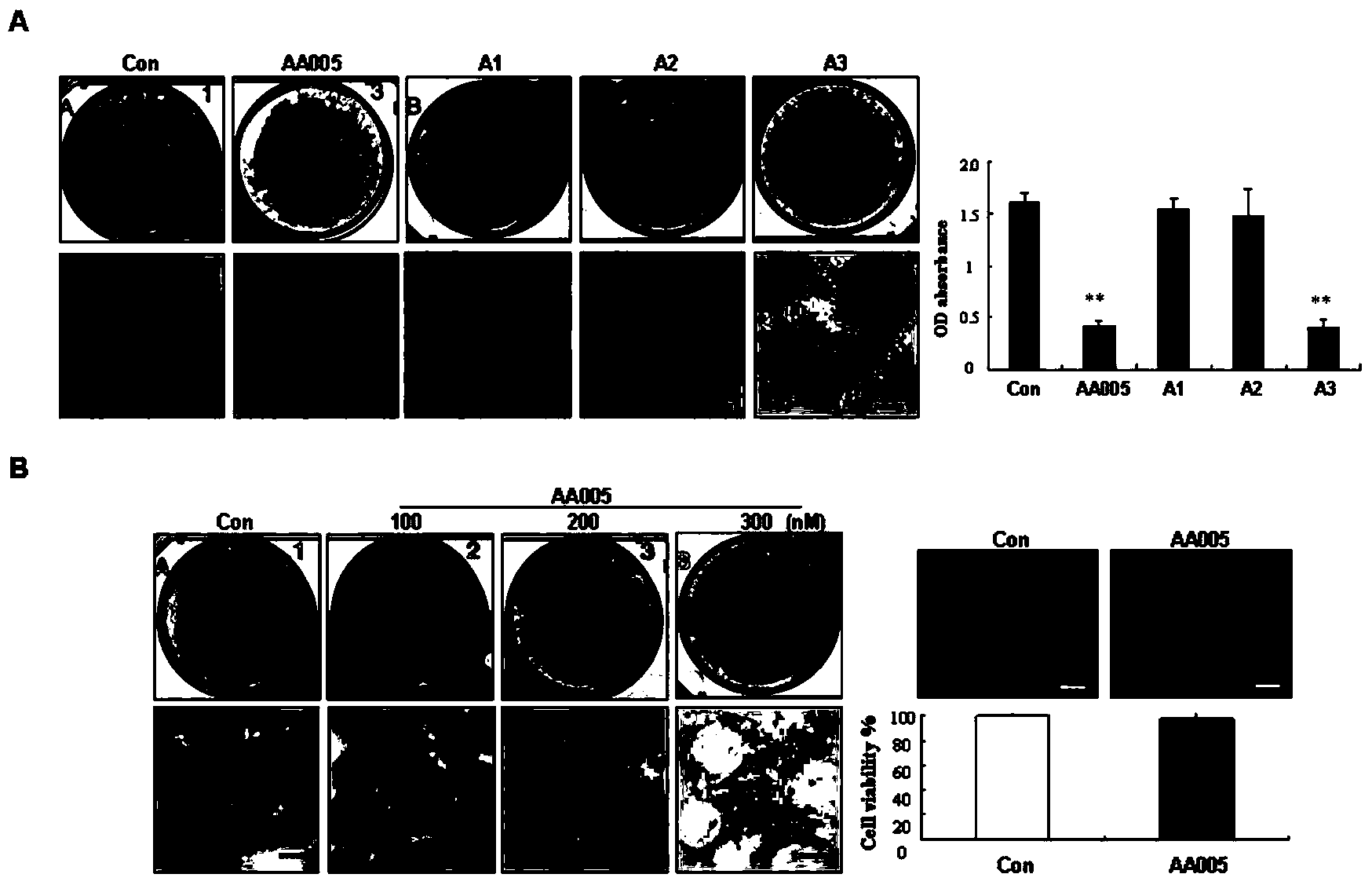
[0051] Example 2. AA005 and its structural analogues can specifically inhibit the accumulation of triglycerides in preadipocyte 3T3-L1
[0052] 3T3-L1 cells were continuously grown to complete contact inhibition for two days, and MDI triple drug was added to DMEM high-glucose medium containing 10% fetal bovine serum to induce differentiation, and 300nM AA005 and structural analogues A1, A2, and A3 were added to the cells in the treatment group , after oil red O staining of cells in the control group and the four compound-treated groups induced to differentiate for 8 days, the macroscopic and microscopic photographs were taken ( image 3 A), oil red O dye in cells was extracted and quantified by spectrophotometer. The results suggested that compared with the control group, the oil red O staining in the AA005 treatment group and the A3 treatment group cells was reduced, and the inhibitory effect of AA005 was the most significant, while the oil red O staining in the A1 and A2 tre...
Embodiment 3
[0053] Example 3. Chemical proteomics fishing for interacting proteins of AA005
[0054] In order to obtain AA005-interacting proteins by affinity chromatography, we connected Biotin outside its active center to construct biotin-labeled AA005 (Biotin-AA005)( Figure 4 A). The modified small molecule still maintained the biological activity of AA005 in inhibiting lipid accumulation in 3T3-L1 cells, and could significantly inhibit the formation of intracellular lipid droplets ( Figure 4 B). We also analyzed the localization of Biotin-AA005 in 3T3-L1 cells by immunofluorescence staining experiment of Mito-tracker labeled mitochondria and FITC-labeled streptavidin co-staining, the results showed that Biotin-AA005 was almost all localized in mitochondria ( Figure 4 C). Accordingly, we speculate that the interacting protein of AA005 is likely to be located in mitochondria. Next, the mitochondria, nucleus and cytoplasm components of 3T3-L1 cells were separated and purified by d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com