Method for predicating bovine serum albumin-water distribution coefficient of organic compound on basis of molecular structures and model establishment method
A technology of bovine serum albumin and organic compounds, which is applied in the fields of electrical digital data processing, special data processing applications, instruments, etc., can solve problems such as the inability to predict environmental organic compounds, low model prediction performance, and limited types of compounds, etc., to achieve Strong practical application ability, convenient and fast method, and easy to obtain results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
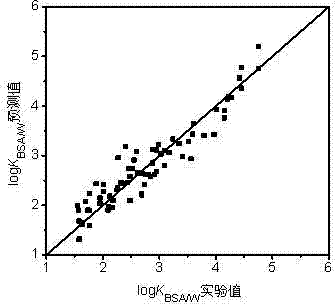
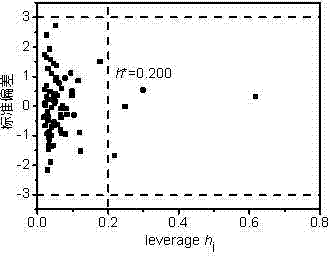
[0030] n-octane: calculated by Williams graph method h i Value is 0.065h *(warning value)=0.200, standard residual ( SE ) = -0.284 > -3, indicating that this compound is within the application domain of the QSAR model. Using the PM6 algorithm of MOPAC 2012, ChemOffice 2010 and EPI Suite calculated 5 descriptors respectively.
[0031] log of n-octane K BSA / W The experimentally determined value was 4.01 [L / kg]. The prediction steps based on the QSAR model are as follows:
[0032] log K BSA / W = 0.747×log K ow + 0.476× E HOMO - 0.024× CCR + 0.870× q + + 0.007× CSEV + 4.342
[0033] = 0.747×(5.81) + 0.476×(-10.434) - 0.024×(19.173) + 0.870×(0.078) +
[0034] 0.007×(145.454) + 4.342
[0035] = 3.87.
Embodiment 2
[0037] Tetrachlorethylene: calculated by Williams graph method h i Value is 0.051h *(warning value)=0.200, standard residual ( SE ) = 2.732 < 3, indicating that this compound is within the application domain of the QSAR model. Using the PM6 algorithm of MOPAC 2012, ChemOffice 2010 and EPI Suite calculated 5 descriptors respectively.
[0038] log of tetrachlorethylene K BSA / W The experimentally determined value was 2.40 [L / kg]. The prediction steps based on the QSAR model are as follows:
[0039] log K BSA / W = 0.747×log K ow + 0.476× E HOMO - 0.024× CCR + 0.870× q + + 0.007× CSEV + 4.342
[0040] = 0.747×(3.40) + 0.476×(-9.545) - 0.024×(-6.035) + 0.870×(0.029) +
[0041] 0.007×(90.284) + 4.342
[0042] = 3.14.
Embodiment 3
[0044] Pyrene: calculated by Williams graph method hi Value is 0.248> h *(warning value)=0.200, standard residual ( SE ) = -0.016 > -3, indicating that this compound is in the application domain of the QSAR model, and the model has good generalization ability. Using the PM6 algorithm of MOPAC 2012, ChemOffice 2010 and EPI Suite calculated 5 descriptors respectively.
[0045] Pyrene's log K BSA / W The experimentally determined value was 4.76 [L / kg]. The prediction steps based on the QSAR model are as follows:
[0046] log K BSA / W = 0.747×log K ow + 0.476× E HOMO - 0.024× CCR + 0.870× q + + 0.007× CSEV + 4.342
[0047] = 0.747×(4.88) + 0.476×(-8.397) - 0.024×(48.446) + 0.870×(0.854) +
[0048] 0.007×(161.662) + 4.342
[0049] = 4.70.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com