Multi-polymerase chain reaction (PCR) primer and method for identifying clouded leopard and application of primer or method in identification of clouded leopard
A multiple, clouded leopard technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of time-consuming, labor-intensive and costly, reduce the difficulty of sampling, economical and practical identification, simple and easy The effect of identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Put 0.5 g of the sample to be tested numbered 3 in Table 1 in a centrifuge tube and cut the sample to be tested in the centrifuge tube with sterilized scissors, then add 500 μL of lysate and 30 μL of 10% ten Sodium dialkyl sulfate and 3 μL of 20 mg / ml proteinase K were thoroughly mixed and then digested in a water bath at 56°C for 12 hours until the liquid in the tube was clear. Add 500 μL of Tris-balanced phenol to the above-mentioned digested mixture, shake slightly for 5 minutes, then place it in a centrifuge at 11000 r / min for 10 minutes, and take the supernatant after centrifugation. Repeat the above centrifugation step twice. Add 1,000 μL of frozen absolute ethanol to the supernatant obtained after repeated centrifugation, place it at -20°C for 1 hour, then centrifuge it in a centrifuge at 12,000 r / min for 13 minutes, discard the supernatant, and add 800 μL of 70% ethanol was shaken slightly for 0.5 min, then placed in a centrifuge at 13000 r / min for 13 min, and ...
Embodiment 2
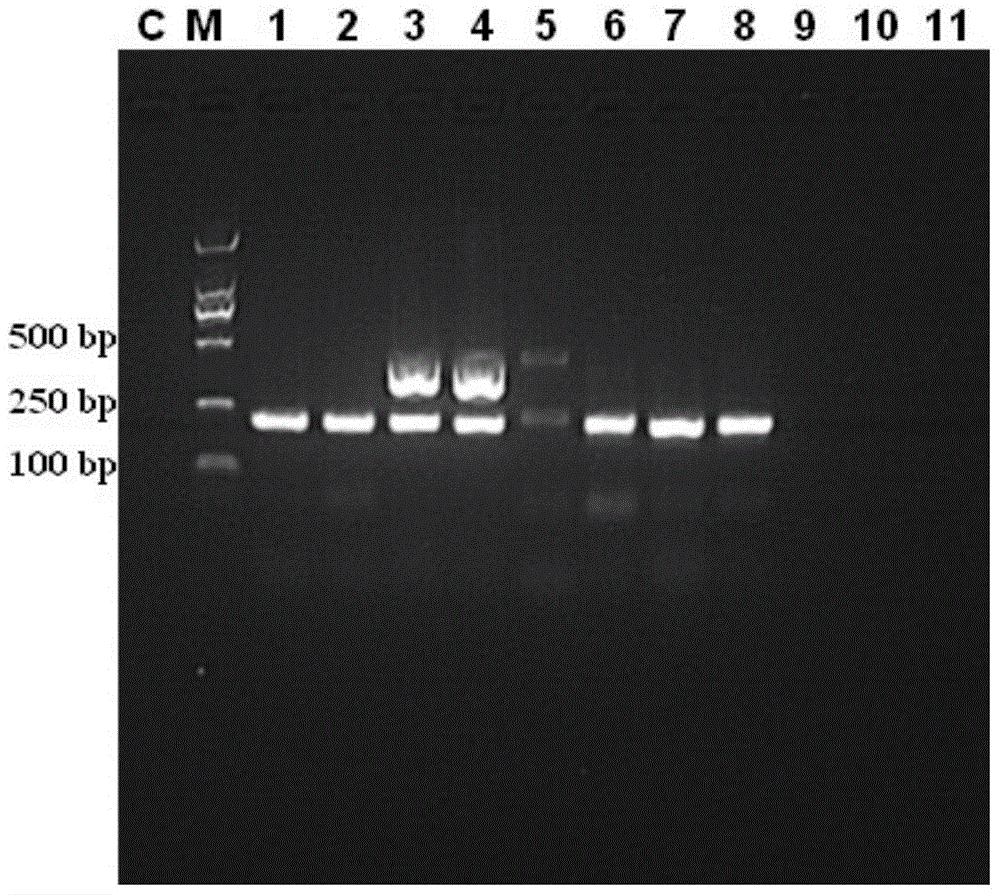
[0038] Operate according to the method of Example 1, the difference is that the sample number to be tested is 4 (in Table 1), the consumption of each primer is 0.75 μ L, the consumption of the total DNA is 45 ng, and the annealing temperature is 55°C, the annealing time is 40s, the electrophoresis results are as follows figure 1 Shown (lane number is 4).
Embodiment 3
[0040] Operate according to the method of Example 1, the difference is that the sample number to be tested is 5 (in Table 1), the consumption of each primer is 1.25 μ L, the consumption of the total DNA is 55 ng, and the annealing temperature is 60°C, the annealing time is 25s, the electrophoresis results are as follows figure 1 Shown (lane number is 5).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com