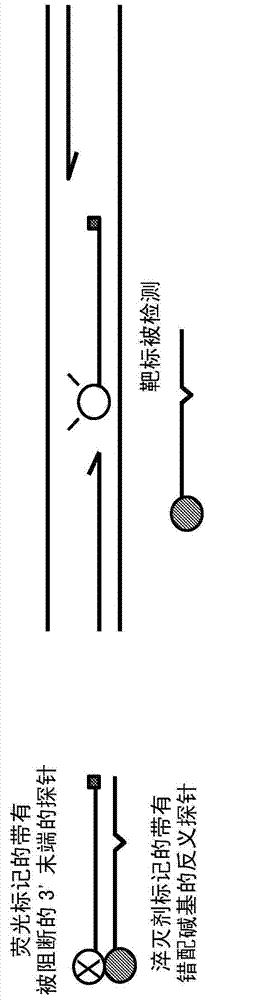
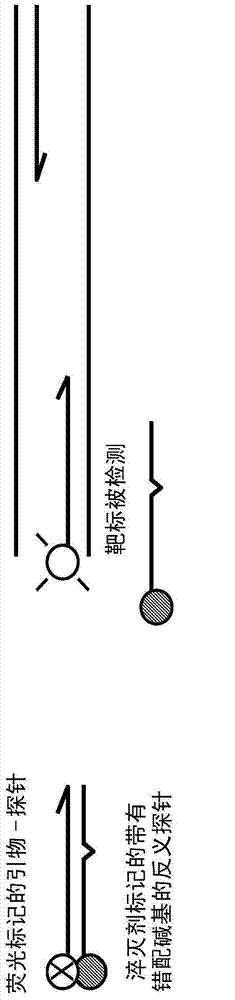
Probe: antiprobe compositions for high specificity dna or rna detection
An antisense probe and probe technology, applied in DNA/RNA fragments, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of expensive targets, complicated washing steps, and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0186] Cycling conditions: Real-time PCR was performed in MX4000 instrument (Stratagene, Inc) with HotStart-IT probe. RTM Quantitative PCR Master Mix (USB, Inc.) (2x) was supplemented with 1 μl of 25 mM MgCl in a 20 tl reaction 2 . To initiate hot-start conditions, tubes were heated at 95°C for 5 min, followed by 40 cycles of two-step PCR (denaturation: 95°C, 15 s; annealing / extension: 58°C, 1 min). The templates used were ULTRAMER.RTM oligonucleotides (DNA Integration Technologies, Iowa, USA) containing EGFR gene fragments targeting EGFR gene fragments with or without T>G transversions, respectively.
example 2
[0188] Internal DDS (iDDS) probe for detection of VKORC1 SNP variants: Antisense probe composition: A significant warfarin dose-related SNP variant is located within the VKORC1 gene encoding vitamin K epoxide reductase subunit 1, At its position 1639, it includes a G>A change relative to the wild-type mutation. To detect these two SNP variants by real-time PCR, the probe, antisense probe and the following primers were used at the following final concentrations:
[0189] VK-1639G-probe: 5'-FAM-cgcacccggccaatg-Phos-3' (SEQ ID NO.: 1) at 200 nM;
[0190] VK-1639G-antisense probe: 5'-catcggccgggtgcg-BHQ1-3' (SEQ ID NO.: 2) at 400nM
[0191] VK-1639A probe: 5'-FAM-attggccaggtgcg-Phos-3' (SEQ ID NO.: 3) at 200 nM
[0192] VK-1639A-antisense probe: 5'-cgcacctggcctat-BHQ1-3' (SEQ ID NO.: 4) at 400 nM
[0193] VK-forward primer: 5'-cctctgggaagtcaagcaag-3' (SEQ ID NO.: 5) at 200 nM
[0194] VK-reverse primer: 5'-aaatgctaggattataggcgtga-3' (SEQ ID NO.: 6) at 200 nM
[0195] Whereas ...
example 3
[0213] In-house DDS (iDDS) probe for real-time PCR detection of single-base variations in the EGFR gene (exon 21L858R mutation) associated with lung cancer diagnosis and treatment: To detect EGFR exon 21 suspected to be present in nucleic acid samples In the presence of mutated codons L858R, and possibly 858L wild-type (normal) codon sequences, the following oligonucleotide probes, antisense probes and PCR primers were synthesized and used at the indicated final concentrations:
[0214] EGFR858R probe: FAM-cagattttggccgggccaaactg-phos (SEQ ID NO.: 7) at 200 nM
[0215] EGFR858R antisense probe: cagtttggcccgcccaatatctg-BHQ1 (SEQ ID NO.: 8) at 400 nM
[0216] EGFR858L probe: CalRed610-cagattttgggctgaccaaactg-phos (SEQ ID NO.9) at 200nM
[0217] EGFR858L antisense probe: cagtttggccagcccataatctg-BHQ2 (SEQ ID NO.: 10) at 400 nM
[0218] Forward primer: gaaaacaccgcagcatgtC (SEQ ID NO.: 11) at 200 nM
[0219] Reverse primer: ctgcatggtattctttctcttcc (SEQ ID NO.: 12) at 200 nM
[0...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com