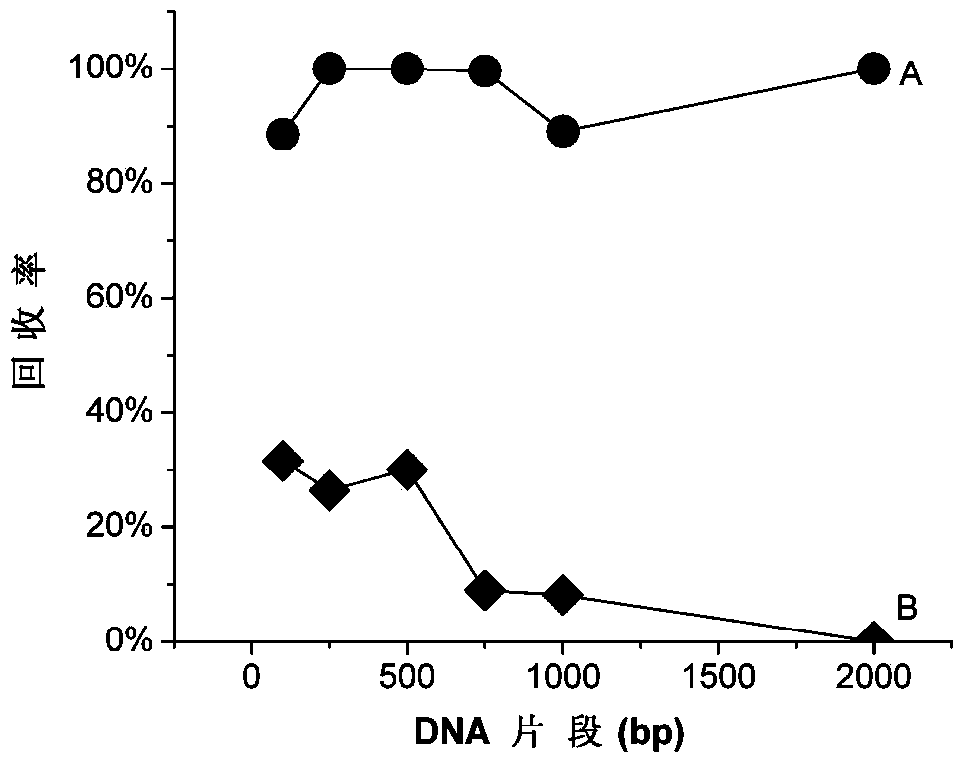
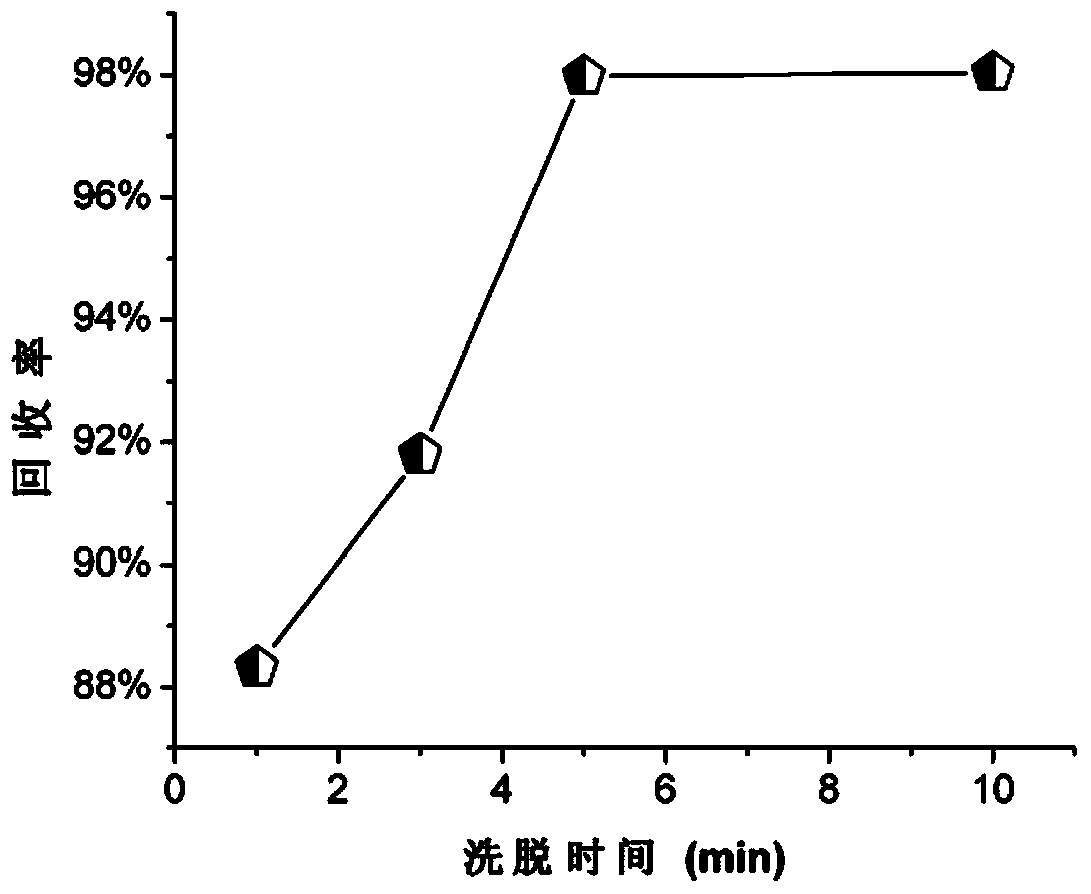
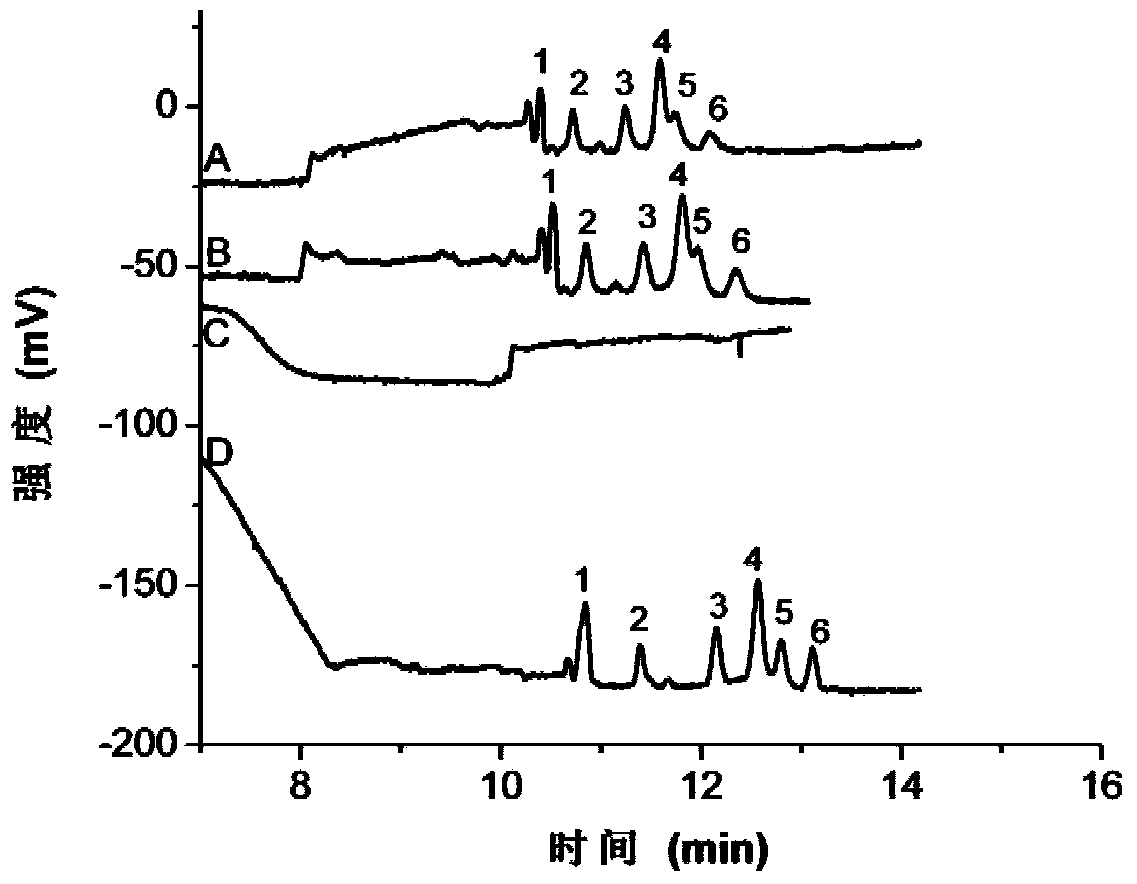
DNA (desoxyribonucleic acid) eluent and elution method
A deoxyribonucleic acid and eluent technology, which is applied in the field of separation and purification of deoxyribonucleic acid, can solve the problems of inability to effectively overcome gravity and low elution capacity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Deoxyribonucleic acid (DNA) adsorption: 3 microliters of DL2000 DNA Marker with a concentration of 90μg / mL and 5 microliters of 4g / L dendrimer-modified silica nanomaterial (SNP-PAMAM) as solid-phase adsorption materials Add a 200 μl centrifuge tube, shake for 10 min at room temperature, stand for 10 min at 4° C., centrifuge at 3500 rpm for 10 min, and collect the SNP-PAMAM-DNA complex.
[0035] The residual DNA content in the supernatant was detected with a fluorescence spectrophotometer (Cary Eclipse, Varian, Palo Alto, CA, USA) using ethidium bromide as a fluorescent probe. The specific method is as follows:
[0036] Add 1.5 microliters of 1mg / mL ethidium bromide (EB) solution to a 5mL graduated test tube, add three times distilled water to make up to the mark, shake up, and excite at 510nm wavelength on a fluorescence spectrophotometer and emit at 620nm wavelength ( The emission and excitation slits are both 10nm, the thickness of the fluorescent cell is 1cm), and the fl...
Embodiment 2
[0043] Add 4 microliters of herring sperm DNA (100μg / mL) and 5 microliters of 4g / L dendrimer modified silica nanomaterials (SNP-PAMAM) as solid-phase adsorption materials into a 200 microliter centrifuge tube, and shake at room temperature After 10 minutes, centrifuge for 10 minutes at 3500 rpm to collect the SNP-PAMAM-DNA complex.
[0044] The residual DNA content in the supernatant was detected with a fluorescence spectrophotometer (Cary Eclipse, Varian, Palo Alto, CA, USA) using ethidium bromide as a fluorescent probe, and the amount of DNA bound to the solid-phase adsorbent was calculated.
[0045] Add the collected SNP-PAMAM-DNA complex to a centrifuge tube containing 15 microliters of eluent. The eluent contains buffer components of 1mM phosphoric acid, 3mM sodium hydroxide and 2mM sodium tetraborate and 5mM [ C 8 MIM]BF 4 Ionic liquid, and the pH of the eluent is 10.8. After shaking for some time at room temperature, centrifuge at 3500 rpm for 10 min, collect the supernatan...
Embodiment 3
[0048] The DNA used in this embodiment is DL2000 DNA Marker (with a concentration of 90 μg / mL). Unless otherwise specified, the operating parameters of this embodiment are the same as those of Embodiment 1.
[0049] The DNA adsorption process uses 3 microliters of DL2000 DNA Marker (with a concentration of 90 μg / mL), and undergoes adsorption, elution, and separation and detection by capillary electrophoresis-UV technology.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com