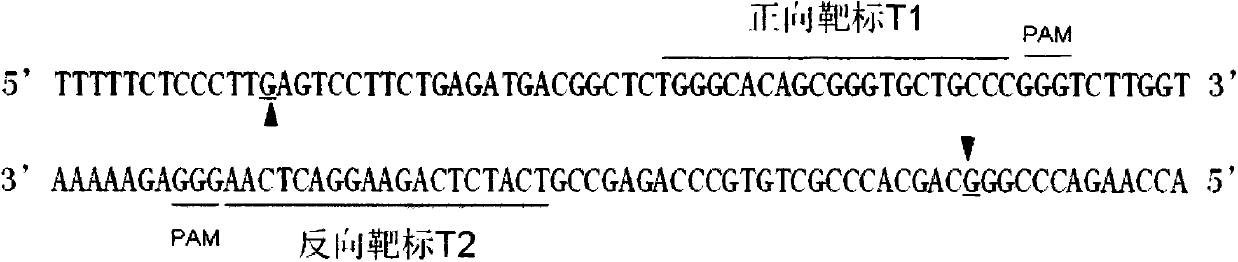Pair of small guide RNAs (Ribonucleic Acids) (sgRNAs) for specifically identifying sheep DKK1 gene and coded DNA (Deoxyribonucleic Acid) and application of sgRNAs
A technology that specifically recognizes, DNA molecules
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Embodiment 1, the construction of sgRNA expression plasmid pair
[0021] 1. sgRNA target design
[0022] Extract a sequence of the sheep DKK1 gene from the sequence of NCBI GI: 417531892 (including the coding region of the first exon and its 5' untranslated region, region: 6667599-6667800, as shown below), and input it into the online software Target sites were designed (http: / / zifit.partners.org / ZiFiT).
[0023] 1 cgcggggact aggcgtgcaa agcgacactc ctcctttctc tttttctccc ttgagtcctt
[0024] 61 ctgagatgac ggctctgggc acagcgggtg ctgcccgggt cttggtcgcc ctggtggctg
[0025] 121 cggctctttg cggtcaccct ctgctgggag tgagcgccac cttgaacttc gttctcaact
[0026] 181 ccaacgccat caagaacacc cc
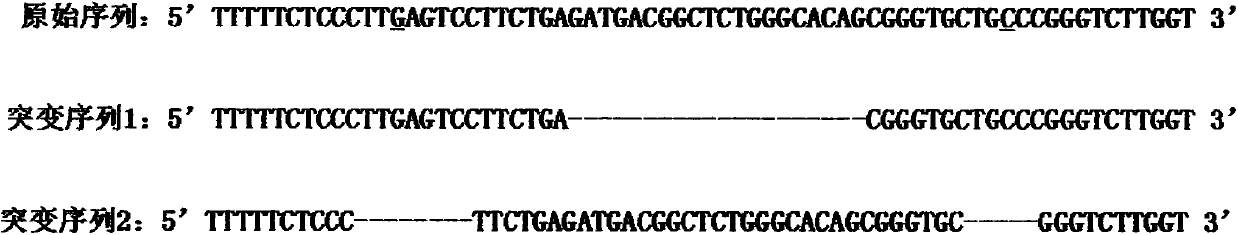
[0027] The forward target sequence is GGGCACAGCGGGTGCTGCCC (77-96) and the reverse target sequence is 5'GTCATCTCAGAAGGACTCAA3' (51-70). The two target sequences are arranged in a "head-to-head" manner, and the distance between the two is 6 bp, that is, there is a 6 bp interval. Name them as forw...
Embodiment 2
[0035] Example 2. Verifying the biological activity of the sgRNA expression plasmid pair constructed in Example 1 by sequencing
[0036] Sheep Fetal Fibroblasts (SEF): For isolation and culture methods, please refer to the literature: Meng Fanhua, Xiao Hongmei, Zhang Dong, Zhou Huanmin. Isolation, culture and characteristic analysis of Mongolian sheep fetal skin fibroblasts. "China Animal Husbandry and Veterinary Medicine" 2012 No. 3, 136- 140.
[0037] 1. Co-transfect 4 μg of each of the recombinant plasmids pX335-sgRNA-F and pX335-sgRNA-R into 1×10 6 SEF cells to obtain recombinant cells. The specific steps of transfection are as follows: use a nuclear transfer instrument (Amaxa, model: AAD-1001S) and a matching mammalian fibroblast transfection kit (Amaxa, product number: VPI-1002) for transfection. First, use 0.05% trypsin (Gibco, catalog number: 610-5300AG) to digest adherent cells, stop digestion with fetal bovine serum (Gibco, catalog number: 16000-044), and wash with...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com