Cloning and Analysis of a 7α,15α Dihydroxylated dhea P450 Enzyme Gene
A DHEA, hydroxylation technology, applied in genetic engineering, oxidoreductase, DNA preparation, etc., can solve the problems of whole-cell catalysis prone to produce by-products, long conversion cycle, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
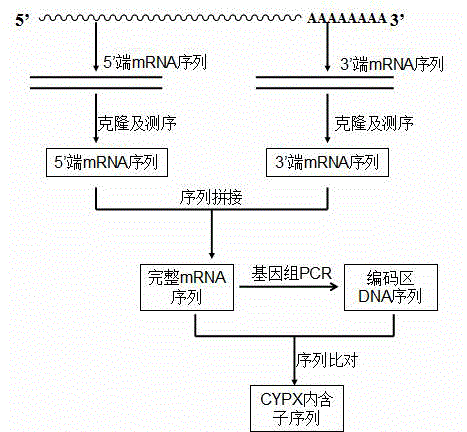
[0032] Embodiment 1 Spirospora flaxensis ( Colletotrichum lini ) Extraction of ST-1 total RNA
[0033] C. liniST-1 was cultured in enzyme-producing medium at 30°C, 220 rpm for 2 days. Collect the bacteria by vacuum filtration, wash 2-3 times with sterile water, put the wet bacteria in a pre-cooled mortar, add 1 mL Trizol reagent, homogenize at low temperature for 2 min; transfer the homogenate to a 1.5 mL centrifuge tube Add 0.2 mL of chloroform, shake vigorously for 30 s, place at room temperature for 3 min, centrifuge at 12,000 rpm at 4 °C for 10 min; transfer the upper aqueous phase to a clean centrifuge tube, add 1 / 2 times absolute ethanol (v / v) , mix well; use a pipette to transfer the mixture to the adsorption column, let it stand at room temperature for 2 minutes, centrifuge at 12,000 rpm for 3 minutes, discard the waste liquid in the collection tube; put the adsorption column back into the collection tube, add 500 μL RPE solution, let it stand for 2 minutes, centri...
Embodiment 2
[0034] Embodiment 2 Spirospora flaxensis ( Colletotrichum lini ) Cloning of ST-1 3' mRNA sequence
[0035] by C. lini ST-1 total RNA was used as a template, and AMV reverse transcriptase was used to perform reverse transcription reaction to synthesize the first strand of cDNA. The reverse transcription reaction was carried out according to the following conditions: incubation at 42-60°C for 15-30 minutes, inactivation of AMV reverse transcriptase at 99°C for 5 minutes, and storage at 5°C for 5 minutes.
[0036] In the reverse transcription tube add Ex Taq For HS enzyme, perform the first round of PCR with M13 Primer M4 and FP1 as primers at the same time. The reaction conditions are 94°C for 2 min, 30 cycles (94°C for 30s, 52°C for 30s, 72°C for 45s), and 72°C for 10min. The PCR product was analyzed by 2% agarose gel electrophoresis, the target band was recovered and ligated with PMD19-T (PMD19-T-CYPX), transformed into JM109, and sent to Shanghai Sangon for sequencing a...
Embodiment 3
[0037] Embodiment 3 Spirospora flaxensis ( Colletotrichum lini ) Cloning of ST-1 5' end mRNA sequence
[0038] The first round of amplification was performed with the 5'-end RACE Outer Primer and RP1 as primers, and the reaction conditions were: 94°C for 3 min, 30 cycles (94°C for 30 s, 65°C for 30 s, 72°C for 90 s), 72°C for 10 min ; Use Inner Primer and RP2 as primers for the second round of PCR. The reaction conditions are: 94°C for 3 min, 30 cycles (94°C for 30s, 55°C for 30s, 72°C for 90s, and 72°C for 10 min. The two rounds of PCR products Analyzed by 1% agarose gel electrophoresis, the target band was recovered by tapping the gel and ligated with PMD19-T (PMD19-T-CYPX5'), transformed into JM109, and then sent to Shanghai Synergy for sequencing.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com