Paeonia lactiflora 2-C-methyl-D-erythritol-4-cyclodiphaophate synthase (PLIspF) gene, and coded product and application thereof
A technology of cyclic diphosphate synthase and bad diphosphate synthase, which is applied in the field of biological and medicinal plant genetic engineering to achieve the effect of increasing the content of terpenoids
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1. Construction of peony cDNA library
[0019] 1. Isolation and detection of total RNA of peony
[0020] Take 2g of the root of Paeonia lactiflora, quickly grind it into powder with liquid nitrogen in a mortar, and quickly transfer it to 65℃ preheated 10mL extraction buffer (CTAB(W / V)2%, Tris-HCl(pH8) .0)100mmol·L -1 , EDTA 25mmol·L -1 , NaCl 2.0mol·L -1 , PVP402%, spermidine 0.5g / L, mercaptoethanol 2%), shake and mix thoroughly; extract twice with equal volume of chloroform, centrifuge at 7500g for 15 minutes. Add 1 / 4 volume of 10M LiCl to the supernatant, mix well and place it for precipitation overnight at 4°C; centrifuge at 7500g for 20 minutes, and use 500μL SSTE (SDS 0.5%, NaCl 1mol·L) for precipitation -1 , Tris-HCl (pH8.0) 10mmol·L -1 , EDTA 1mmol·L -1 , Dissolve at 65°C for 5 minutes. Extract with an equal volume of chloroform, centrifuge at 13000g for 5 minutes; add 2 volumes of absolute ethanol to the supernatant, and place at -70°C for 2 hours; centrifug...
Embodiment 2
[0023] Example 2: Cloning of peony related genes
[0024] Randomly pick 5000 single clones for colony PCR identification. Take an appropriate amount of PCR thin-walled tubes, place them on ice, and add 17.3ul of sterilized water to each tube. Use a sterilized 10ul small pipette tip to pick up the single leukoplakia into sterilized water, shake and mix. Add sequentially: Taq buffer 2.5μL, MgCl 2 (25mM) 1.8μL, dNTP (2.5mM) 1μL, M13+ primer (10pmol) 1μL, M13-primer (10pmol) 1μL, Taq enzyme 0.4μL. PCR reaction conditions were 94°C pre-denaturation for 5 minutes, 94°C for 40 seconds, 54°C for 40 seconds, 72°C for 4 minutes, after 35 cycles, 72°C extension for 10 minutes, and 4°C storage. After the PCR reaction enters 4°C, remove the PCR thin-walled tube, take 7ul PCR product and add 3ul bromofin for agarose gel electrophoresis, take a picture after half an hour, observe the gel image, and roughly identify the size and size of the insert based on the gel image Fragment rate. The si...
Embodiment 3
[0025] Example 3 Bioinformatics analysis of PLIspF gene
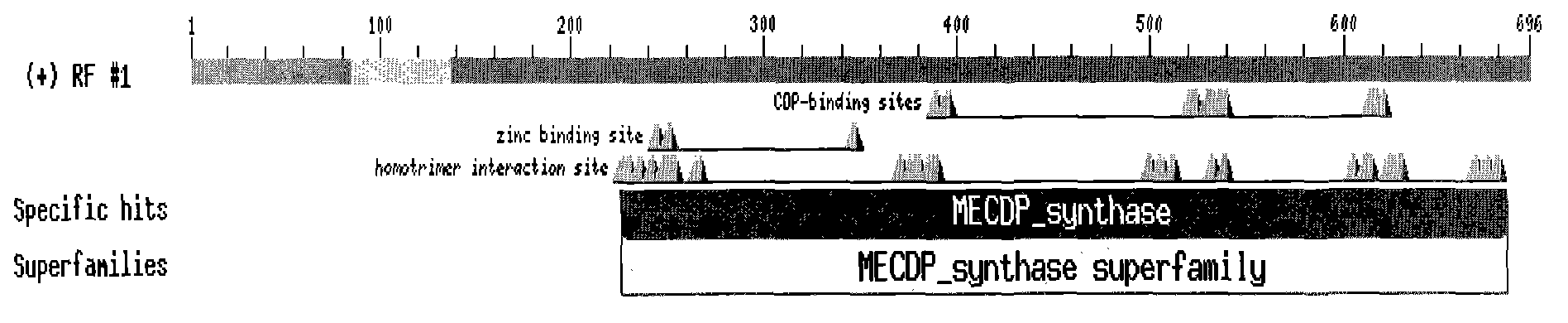
[0026] The length of the full-length cDNA of the Paeonia lactiflora 2-methyl-D-erythritol-2,4-cyclodiphosphate synthase gene involved in the present invention is 696 bp, and the detailed sequence is shown in sequence 1 in the sequence table, where the open reading frame is located 1-696bp. The full-length cDNA sequence of Paeonia lactiflora was searched for nucleotide homology in Non-redundant GenBank+EMBL+DDBJ+PDB and Non-redundant GenBank CDS translation+PDB+Swissprot+Superdate+PIR database using BLAST program. The gene has high homology with IspF in other species at the amino acid level, and has a typical MECDP_synthase (2-C-methyl-D-erythritol-2, 4-cyclodiphosphate synthase, IspF) domain. Such as figure 1 .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com