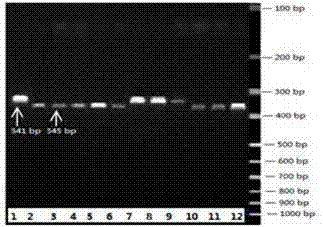
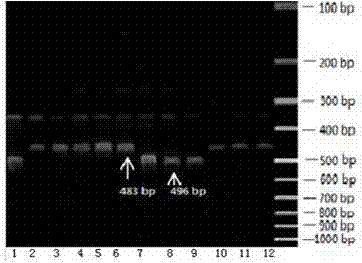
Bilateral molecular marker identification technology of melon wilt
A technology of melon wilt and molecular markers, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve misjudgment, inaccurate disease resistance of melon wilt, and the influence of temperature and humidity on inoculation and observation time And other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0018] 1. DNA extraction
[0019] DNA was extracted by CTAB method. The DNA extraction effect is better than the SDS extraction method, and the sugar removal effect is obvious. The specific method is as follows:
[0020] (1) Take a small amount of leaves and place them in an Eppendorf tube (about 70-100 mg), and freeze them in liquid nitrogen for 1 min;
[0021] (2) Open the cap of the tube and mash the young leaves with a small wooden stick (note: mash for a while and then put it in liquid nitrogen, then mash it after freezing until it is very broken, generally 3 times); then, put Put it into 3 small steel ball tubes, shake the oscillator 3-4 times;
[0022] (3) Depending on the amount of material, add 600-700 ul of preheated DNA extraction solution (CTAB), mix gently, and place in a 65°C water bath for 25 minutes, shaking 2-3 times during the process;
[0023] (4) Add an equal volume of phenol / chloroform (600-700ul) into the tube, invert it upside down for 5 minutes to m...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com