Heparinase I fusion protein
A technology of fusion protein and heparinase, applied in the field of genetic engineering, can solve problems such as low proportion, large molecular weight, and reduced specific activity of fusion protein
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1. Construction of fusion expression vector
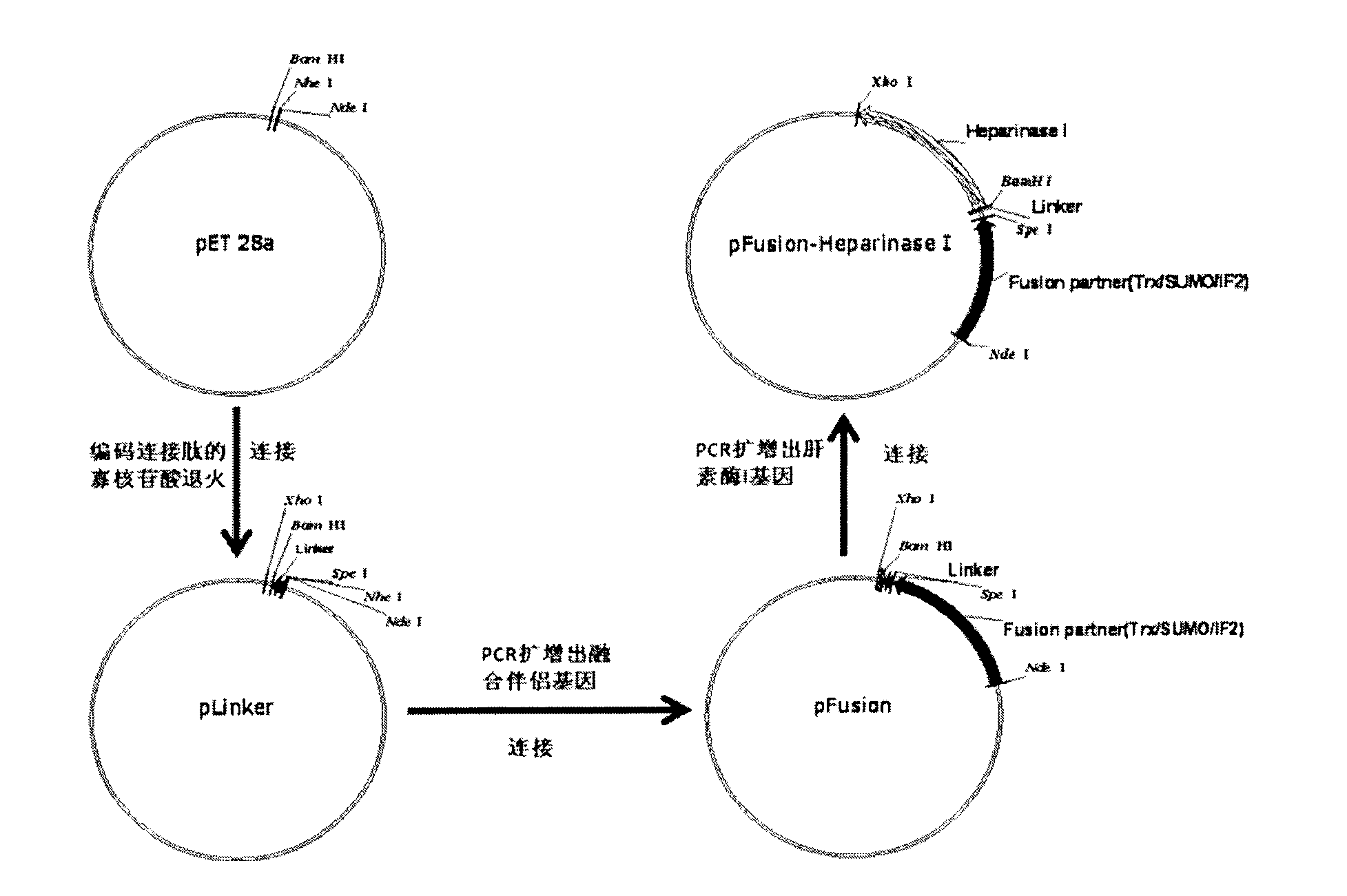
[0035] The construction process of the fusion expression vector pFusion is as follows: figure 1 As shown, the specific process is as follows: two oligonucleotide chains are synthesized, and the sequences are respectively:
[0036] Linker sense (SEQ ID No.1)
[0037] 5' ctagc Actagt ggtggtggtggttctggtggtggtggttctgatgatgatgataaagcgg-3'
[0038] Linker antisense (SEQ ID No.2)
[0039] 5'-gatcccgctttatcatcatcatcagaaccaccaccaccagaaccaccaccacca actaggtg -3'
[0040] 100 pmol oligonucleotide chains were mixed respectively, heated at 70°C for 15 minutes, cooled naturally to room temperature, and annealed to form double strands. The pET 28a vector was digested with Nhe I and BamH I, and recovered from the gel to obtain a linearized vector. The annealed product and the linearized vector were ligated with T4 ligase, and transformed into Escherichia coli DH5α. The recombinant vector was named pLinker. The insert seque...
Embodiment 2
[0054] Example 2. Construction of Flavobacterium heparin heparanase I fusion expression vector
[0055] The construction process of Flavobacterium heparin heparinase I fusion expression vector is as follows: figure 1 As shown, the specific process is as follows: amplify the heparanase I gene from the chromosomal DNA of Flavobacterium heparinus, and the primers are:
[0056] FH F (SEQ ID No. 10): 5'-gatcggatcccagcaaaaaaaatccggtaac-3'
[0057] FH R (SEQ ID No. 11): 5'-gatcctcgagttatctggcagtttcgctgtac-3'
[0058] The PCR reaction system is: 100ng Flavobacterium heparin genomic DNA, 10pmol of each primer, 0.2mM of each dNTP, 1x amplification buffer, 2 units of pfu enzyme. The PCR reaction program was: 94°C for 5 minutes; 30 cycles of 94°C for 30 seconds, 55°C for 30 seconds, 72°C for 2 minutes; 72°C for 5 minutes. The PCR product was detected by 1% agarose gel electrophoresis, and the size was 1092bp.
[0059] The PCR product was double-digested with BamH I and Xho I, and then...
Embodiment 3
[0060] Example 3. Expression of Heparanase I Fusion Protein
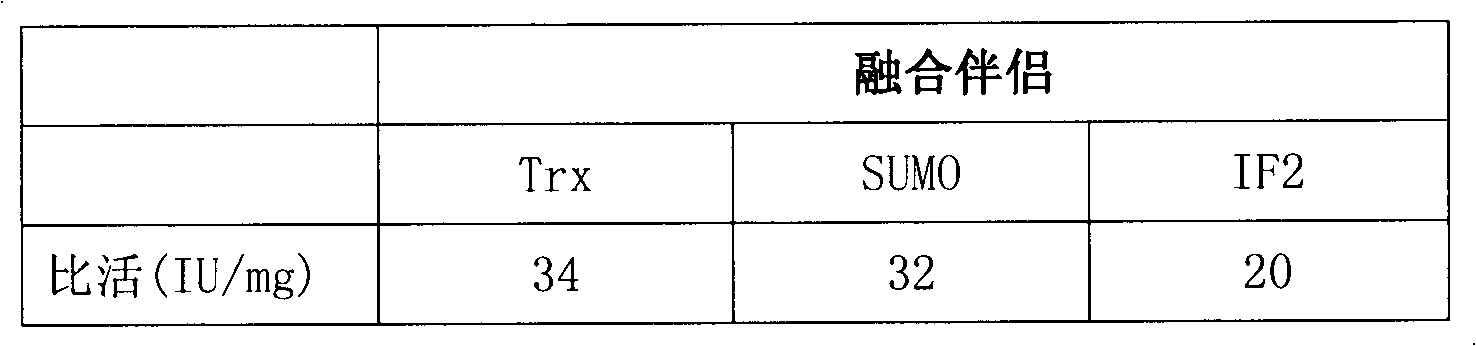
[0061] Transform pTrx-FH Heparinase I, pSUMO-FH Heparinase I, and pIF2-FH Heparinase I into Escherichia coli BL21(DE3) competent cells respectively, and culture them on LB solid medium (containing 10 μg / ml kanamycin) at 37°C overnight. Pick a single colony to 3ml LB medium (containing 10ug / ml kanamycin), culture at 37°C until OD 600 = 0.5, add IPTG with a final concentration of 0.5 mM, and continue culturing at 16° C. for 12 hours. Collect the cells by centrifugation at 10,000 g, wash with 400 μl PBS (50 mM NaH 2 PO 4 pH 8.0, 300mM NaCl), resuspended, and ultrasonicated. Take 200μl of the lysate, centrifuge at 12,000g for 10 minutes, and collect the supernatant; use 200μl of PBS (50mM NaH 2 PO 4 pH 8.0, 300mM NaCl) resuspended. The ultrasonic total protein, ultrasonic supernatant and ultrasonic precipitation were analyzed by 12% SDS-PAGE. The expression result of Flavobacterium heparinus heparanase I fusio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com