Detection method for single nucleotide polymorphism
A single nucleotide polymorphism and detection method technology is applied in the field of molecular biology technology, and can solve the problems of high cost of detection instruments, isotope pollution and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
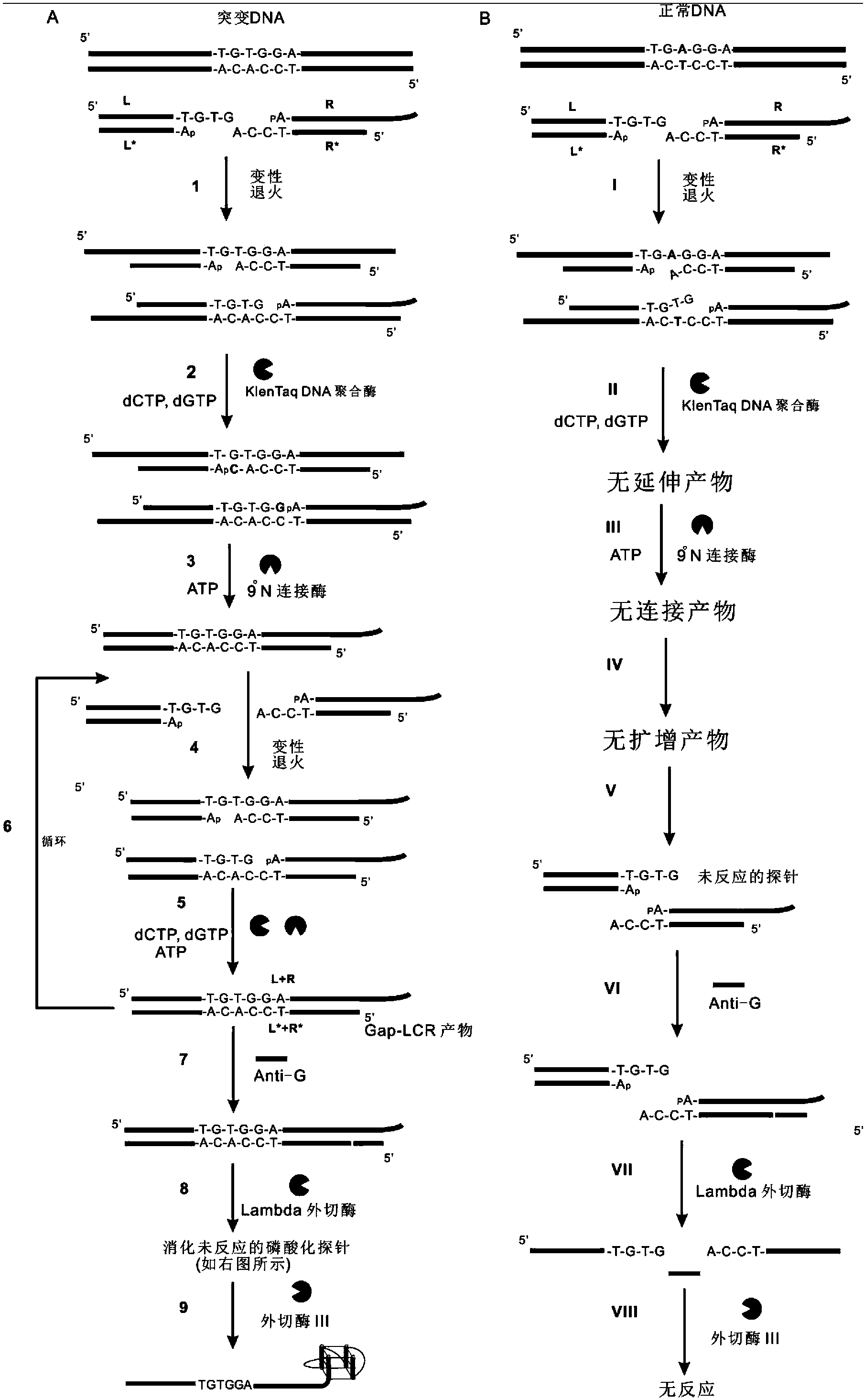
[0038] Example 1. Detection of sickle cell anemia gene DNAT-1 with group A (Probe L, ProbeL*, Probe R, Probe R*) oligonucleotide probes (mutant DNAT-1 is mutant DNA, normal DNAT-1 is normal DNA).
[0039] For the reaction steps to detect the sickle cell anemia gene, see figure 1 . Group A (Probe L, ProbeL*, Probe R, Probe R*) oligonucleotide probes are designed to detect the sickle cell anemia gene, in which the 3' end of the oligonucleotide probe Probe R is labeled with a porphyrin The DNAzyme sequence with peroxidase activity is used for signal detection; the sequence at the 5' end is a sequence that can form a complementarity with the sequence near the mutation site of the target gene to be detected, and is used for Gap-LCR amplification.
[0040] After adding group A oligonucleotide probes Probe L, ProbeL*, Probe R, Probe R* into the Gap-LCR system, under denaturing conditions, the double strand of the DNA T-1 to be tested and the probe (Probe L, ProbeL* , Probe R, Prob...
Embodiment 2
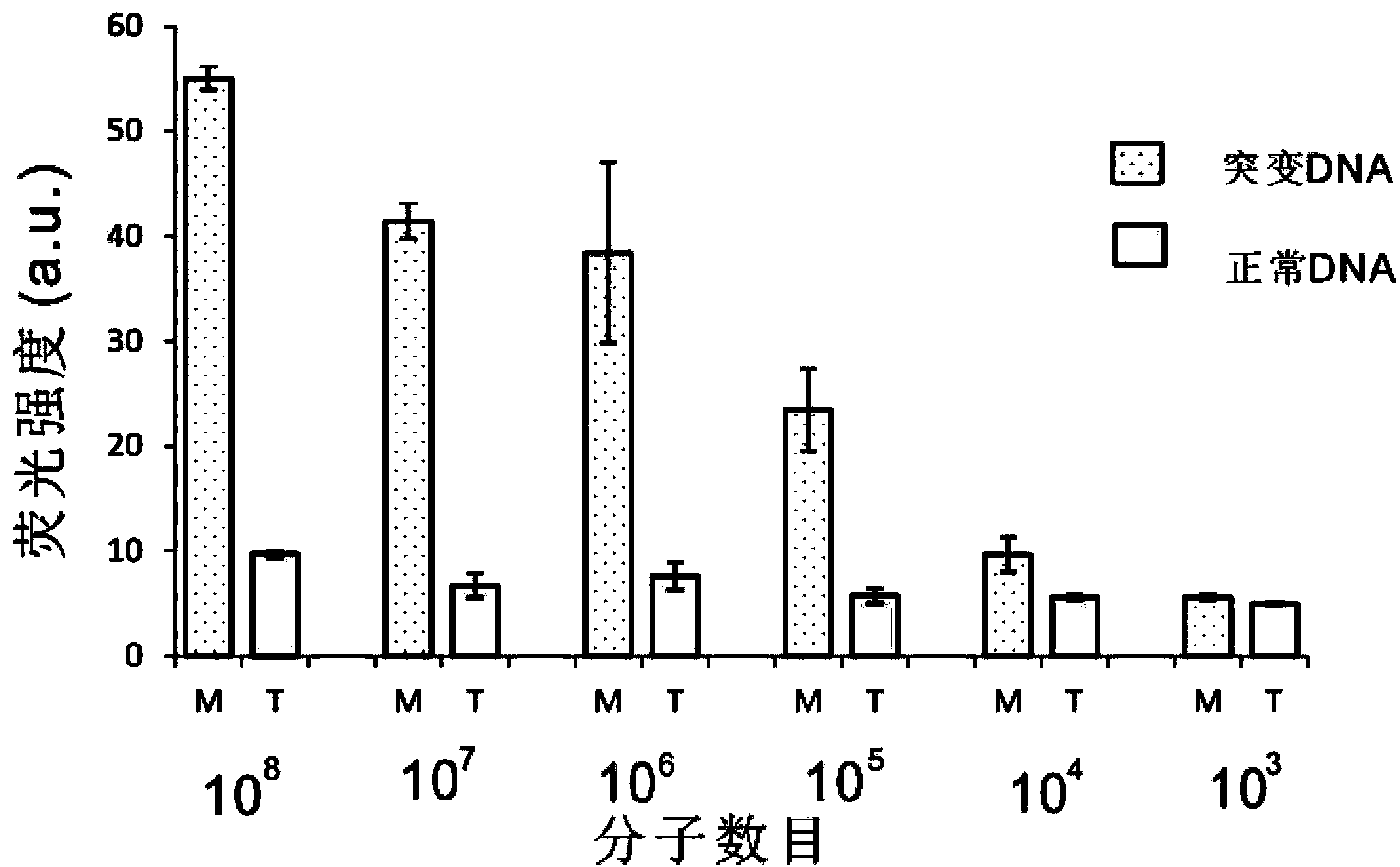
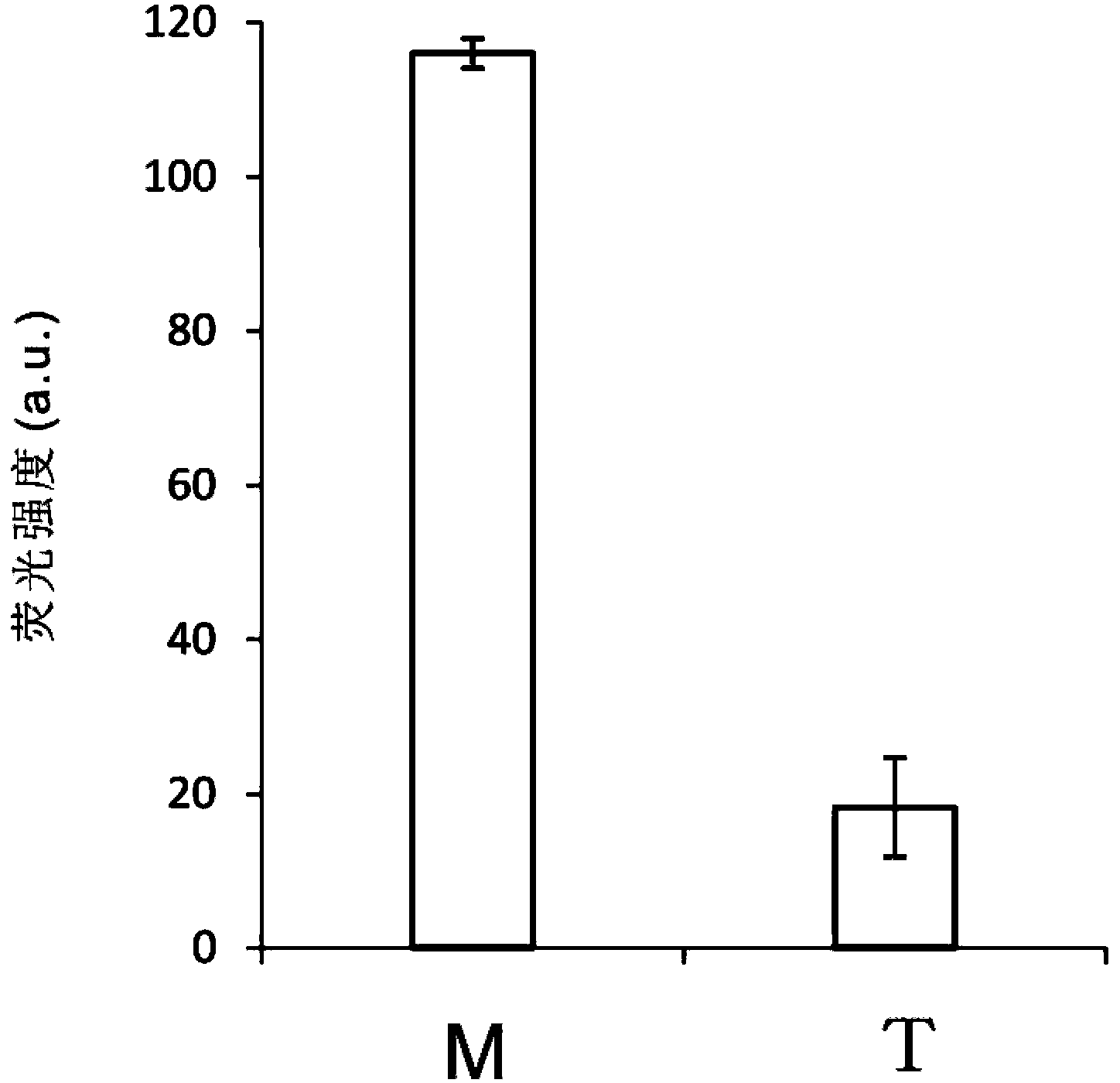
[0058] Example 2. Using group B (B1-B4) oligonucleotide probes to detect the mutation site 235delC in the neonatal deafness gene.
[0059] For the detection results of the mutation site 235delC in the neonatal deafness gene, see image 3 . Group B (B1-B4) oligonucleotide probes are designed to detect the mutation site 235delC in the neonatal deafness gene, in which the 3' end of the oligonucleotide probe B4 is labeled with a compound that can bind to porphyrin iron, etc. The DNAzyme sequence with peroxidase activity is used for signal detection; the sequence at the 5' end is a sequence that can form a complementary sequence with the sequence near the mutation site of the target gene to be detected, and is used for Gap-LCR amplification.
[0060] After adding group B oligonucleotide probes (B1-B4) into the Gap-LCR system, under denaturing conditions, the double-strand structure of the DNA T-1 to be tested and the double-strand structure of probe 1 are opened; Under the condit...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com