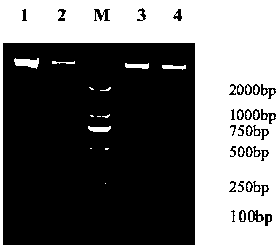
Extraction method for metagenome
A metagenome and extraction method technology, applied in the field of metagenome extraction, can solve problems such as multi-time, and achieve the effects of increasing output, reducing waste of sequencing data, and reducing high background interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Example 1 Probe hybridization subtraction for simulated mixed samples (hybridization first and then coupling)
[0071] 1. Take 20 μl of simulated mixed DNA (according to human whole blood DNA and 2815bp plasmid mixed according to the nanogram amount of 9:1, the total amount is about 100ng / μl) in a 200 μl tube, and place it in a PCR instrument to preheat at 95°C for 5 minutes. 65°C for 5 minutes; take 27 μl of 2×hybridization solution HYB and place it in a PCR instrument to preheat at 65°C for 5 minutes;
[0072] 2. Take 0.1 μl each of the probes whose serial numbers are SEQ ID NO: 1~70, about 70 pmol, and preheat at 65°C for 2 minutes;
[0073] 3. Mix 54 μl of the above system, hybridize at 65°C for 1 hour;
[0074] 4. Take 30 μl of magnetic beads M-280 (invitrogen), wash twice with 2×B&W buffer, discard the supernatant, and resuspend with 54 μl 2×B&W;
[0075] 5. Add the hybridization system (54 μl) to the magnetic beads, mix well, and incubate at room temperature ...
Embodiment 2
[0079] Example 2 Probe hybridization subtraction for total DNA in respiratory lavage fluid samples (hybridization first and then coupling)
[0080] Except in Step 1 of Example 1, select human respiratory lavage fluid sample DNA, take 32 μl of 2×hybridization solution HYB, and take 3 μl (about 120 pmol) of each probe with sequence number SEQ ID NO: 1~4 in Step 2, Except for resuspending with 64 μl 2×B&W in step 4, the same method and conditions as in Example 1 were used to carry out first hybridization followed by coupled probe hybridization subtraction.
Embodiment 3
[0081] Example 3 Probe hybridization subtraction for simulated mixed samples (coupling first and then hybridization)
[0082] 1. Take 30μl magnetic beads M-280 (invitrogen), wash twice with 2×B&W buffer, discard the supernatant, add 15μl 2×B&W buffer to resuspend the magnetic beads;
[0083] 2. Add 0.1 μl (about 70 pmol) and 8 μl of ddH to the resuspended magnetic beads 2 O, incubate at room temperature for 15 minutes, and mix gently during the period; place on a magnetic stand at room temperature for 1 to 2 minutes, and remove the supernatant;
[0084] 3. Take 20 μl of simulated mixed DNA (the human whole blood DNA and about 2.6kbp plasmid are mixed according to the nanogram amount of 9:1, the total amount is about 100ng / μl) in a 200μl PCR tube, and put in the PCR machine to preheat at 95°C for 5 minutes , 65°C for 5 minutes; take 26 μl of 2×hybridization solution HYB and place it in a PCR instrument to preheat at 65°C for 5 minutes;
[0085] 4. Add the mixed DNA and hybr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com