Method for obtaining genome sequence of papaya ringspot virus through high-throughput small RNA sequencing
A virus genome and ringspot virus technology, applied in the fields of virus peptide, genetic engineering, DNA preparation, etc., can solve the problems of economic loss, papaya production and harvest, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] A method for cloning the whole genome sequence of papaya ringspot virus Hainan isolate PRSV-HN-y1 obtained by high-throughput Small RNA sequencing, comprising the following steps:
[0063] (1) Total RNA was extracted from samples infected with papaya mosaic disease, sRNA was separated by polyacrylamide gel electrophoresis (PAGE gel), the sequence information was trimmed after Small RNA sequencing, and the linker sequence was removed to obtain sRNA fragments.
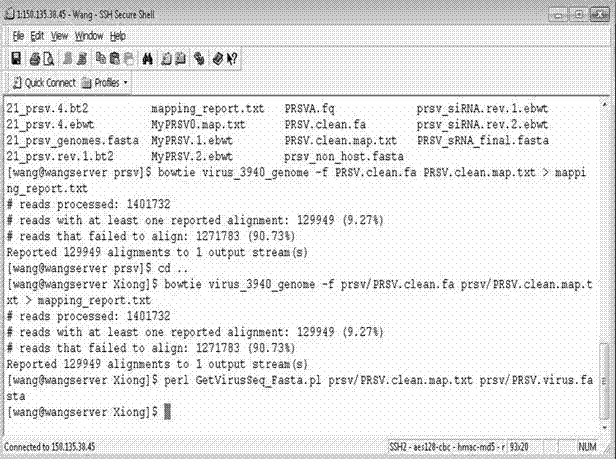
[0064] (2) Compare the sRNA fragments with all known viral genome sequences in Genbank by running Bowtie software (default parameters), and screen out highly matching small RNA fragments; as attached figure 1 shown.
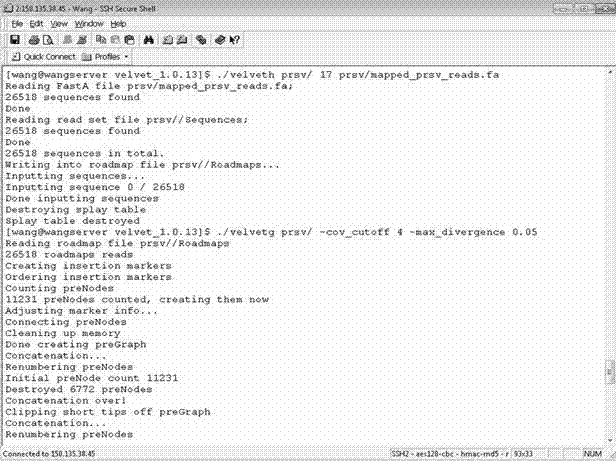
[0065] (3) Run the Velvet software, set the parameters to k-mer=17 and minimum coverage=5, use the small RNA fragments obtained in step 2 as input, and assemble them into short contig sequences; as attached figure 2 , 3 , 4 shown.
[0066] (4) Run the Blast software (default parameters) to compa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com