Oligonucleotide probe, and method for detecting target molecule through using it
An oligonucleotide probe and target molecule technology, which is applied in the field of oligonucleotide probe and warm nucleic acid probe technology to detect target molecules, can solve the problems of high price and complicated operation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
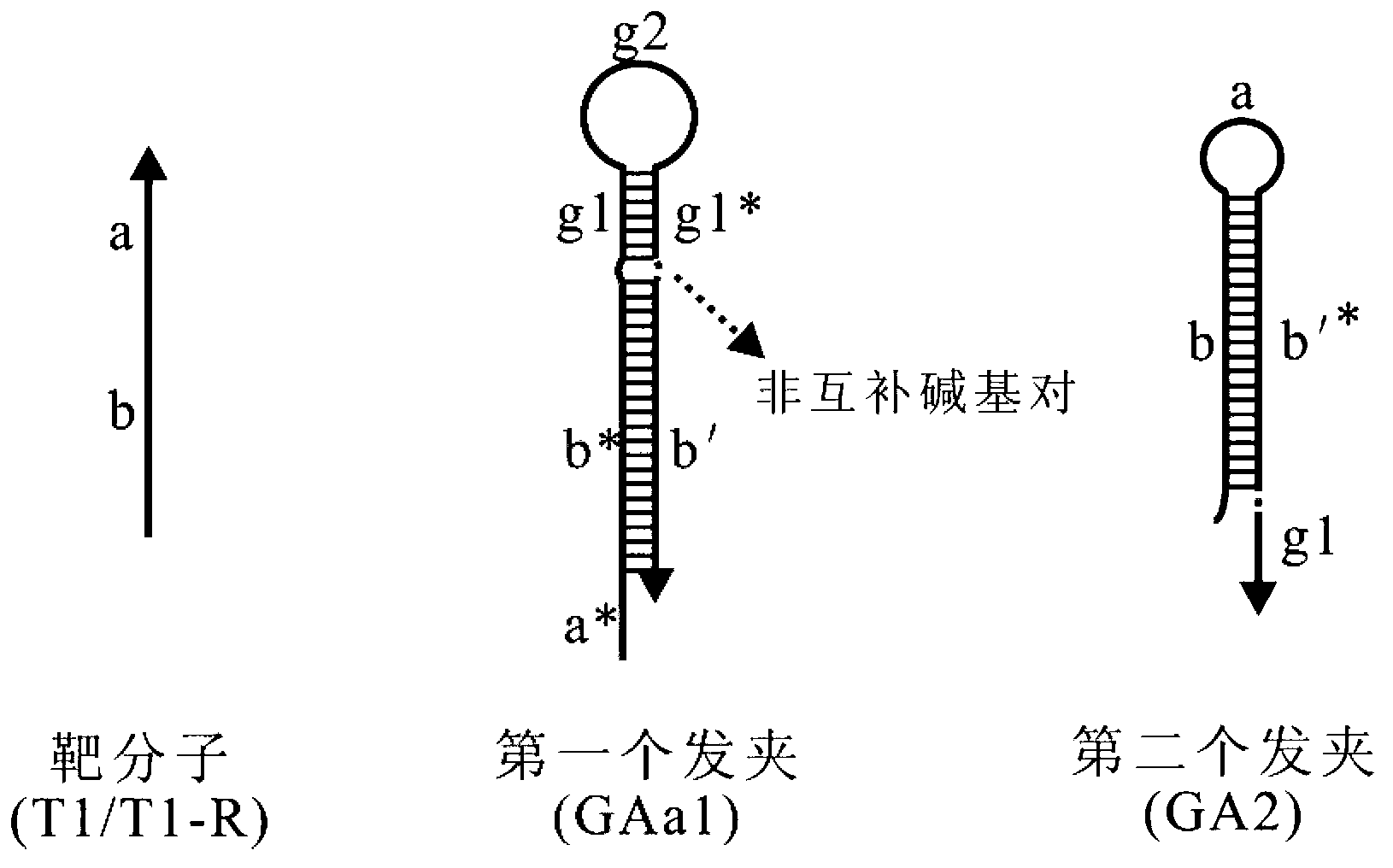
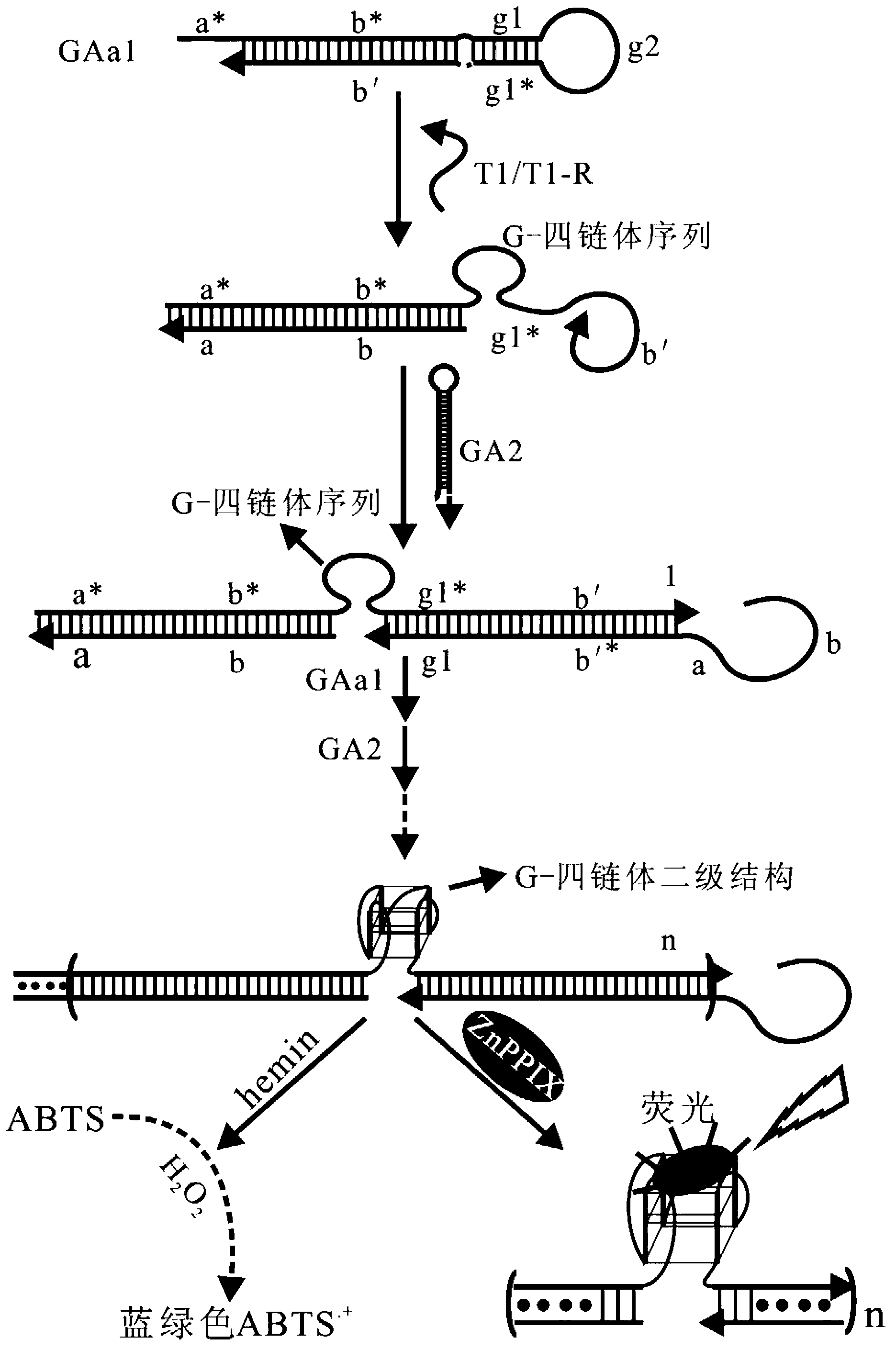
[0061] Embodiment 1, carry out probe design based on an artificially synthesized target nucleic acid molecule (combined with the attached figure 1 )
[0062] Artificially synthesized target nucleic acid DNA sequence T1:
[0063] 5'-TCTCCACAACTGAACACGTTAGACCACTT-3' (SEQ ID NO 2).
[0064] The target nucleic acid RNA sequence T1-R obtained by T1 transcription:
[0065] 5'-UCUCCACAACUGAACACGUUAGACCACUU-3' (SEQ ID NO 3).
[0066] (The double-underscored sequence is named sequence a, and the unmarked part is sequence b.)
[0067] The probe sequences designed according to T1 and T1-R are:
[0068] A. Hairpin GAa1
[0069] 5'- TCTAACGTGTTCAGTTGTGGA T TCCACAACTGAACACGTTAGA-3' (SEQ ID NO 4).
[0070] Sequence composition, relationship between each part and naming instructions:
[0071] ① The double underlined sequence at the 5'-end is a cohesive end, which is complementary to the double-underlined sequence at the 3'-end of the target nucleic acid T1, named sequence ...
Embodiment 2
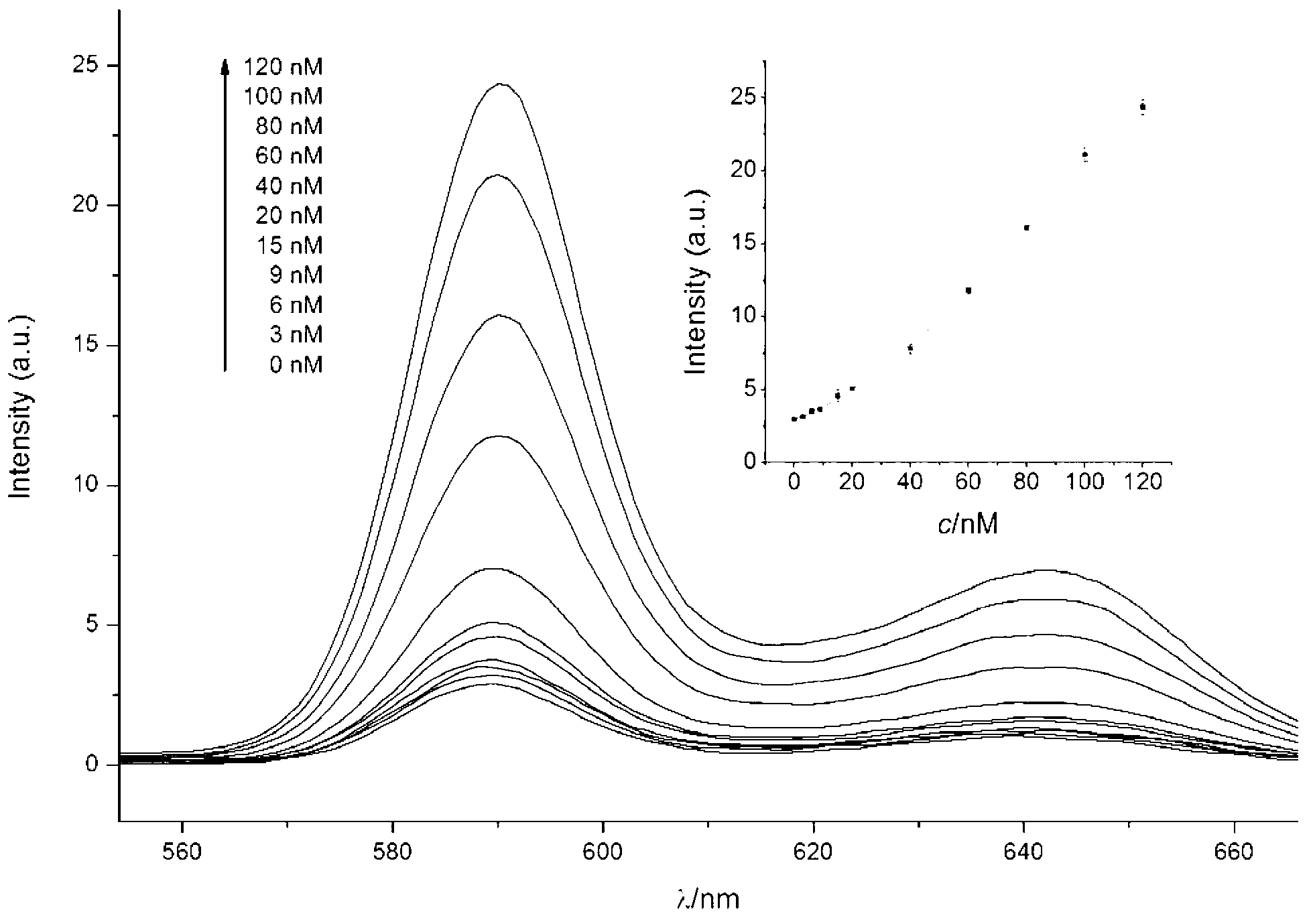
[0080] Embodiment 2, use the probe described in embodiment 1 to detect target nucleic acid
[0081] (1) ABTS chromogenic method detects different concentrations of target nucleic acids
[0082] ①Reaction system (TV=20μL)
[0083] Table 2 Hybrid chain reaction system used in ABTS chromogenic method
[0084]
[0085] Note: X represents the different volumes of target nucleic acid stock solutions added, Y represents the different target nucleic acid final concentrations corresponding to X, respectively 0M, 7.5x10 -9 M,1.5x10 -8 M,3x10 -8 M,5x10 -8 M,7.5x10 -8 M, 1x10 -7 M.
[0086] ②Probe pretreatment
[0087] The probes—hairpins GAa1 and GA2—need to be pretreated before detection. The pretreatment method is: divide the amount of NaCl required for the reaction into two equal parts, and mix it with the hairpin GAa1 and GA2 respectively (taking the amount required for one reaction as an example: 1 μL GAa1 (10 μM) mixed with 1 μL NaCl (4M), 1 μL GA2 (10μM) mixed with 1μ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com