Mutant gene of rice starch branching enzyme SBE3 gene and application of mutant gene
A starch branching enzyme and mutant gene technology, which is applied in the field of molecular genetics, can solve the problems of difficulty in improving the content of resistant starch in rice, being easily affected by the environment, and the complexity of the determination method, so as to achieve accurate and consistent phenotype determination results and speed up Breeding process, the effect of saving breeding costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] (1) Jiangtangdao No.1 / Miryang 23 F 2 Population construction and phenotyping
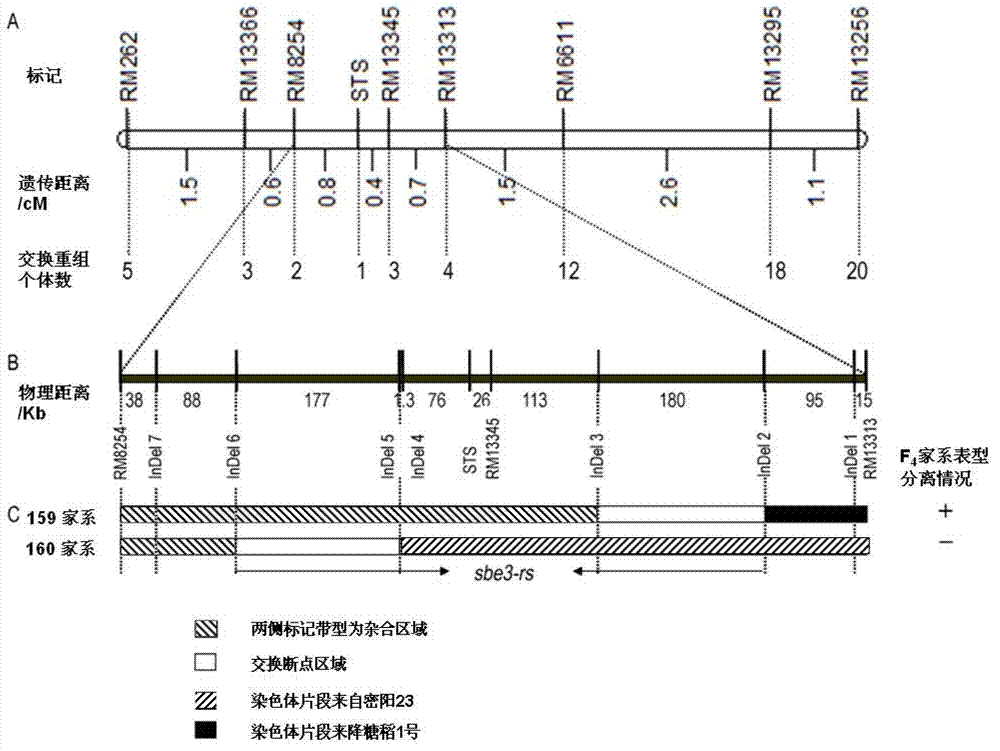
[0045] In the summer of 2008, F 1 generation seeds. The female parent was processed by artificial clipping and emasculation. Hybrid F was planted in Hainan base in the winter of 2008 1 Generation to get F 2 Generation seeds, planted F in Shanghai Academy of Agricultural Sciences in the summer of 2009 2 generation. each F 2 Leaf extraction DNA was collected from individual plants for genotype analysis, and selfed seeds were harvested from individual plants for the determination of resistant starch content. The F2 generation was used to initially map the genes controlling the content of resistant starch, and a QTL related to the content of resistant starch was found in the interval of molecular markers RM13366 and RM6611 on chromosome 2, which could explain 60.4% of the variation in the content of resistant starch. Select heterozygous individual plants in this segment, and further fine-...
Embodiment 2
[0064] Example 2 Validation of Molecular Markers
[0065] 1 Materials and methods
[0066] 1.1 Materials
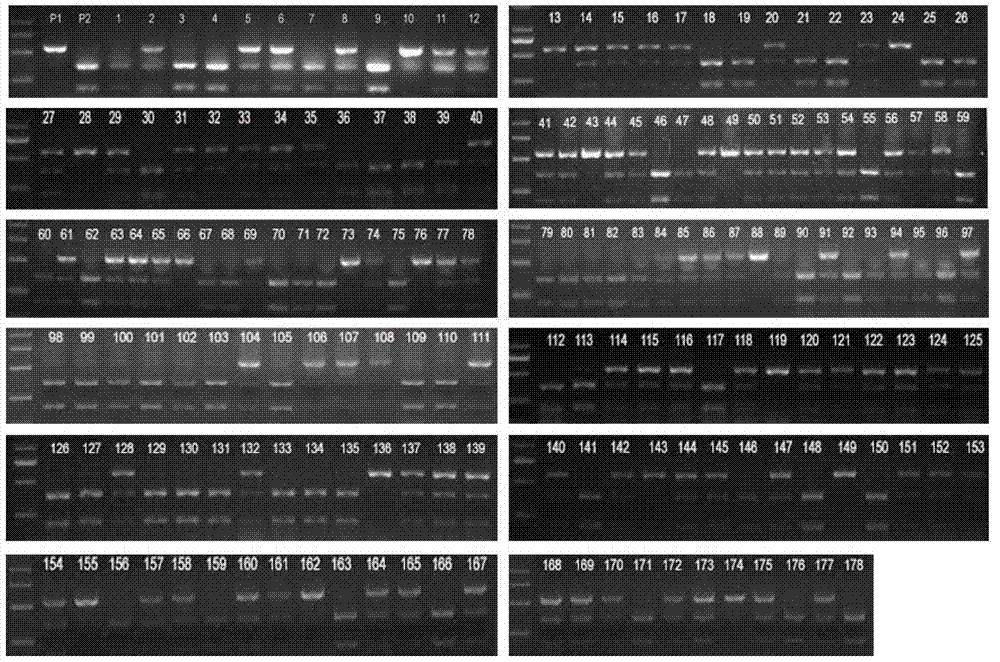
[0067] Jiangtangdao 1 / Miryang 23 Hybrid F 2 Generation of 178 individual plants.
[0068] PCR amplification primers:
[0069] PCR-SpeI F: ATGTGATGTGCTGGATTTGG SEQ ID NO. 3
[0070] PCR-SpeI R: TGTGGTTTTCATACCGTTCTTA SEQ ID NO. 4
[0071] 1.2 Method
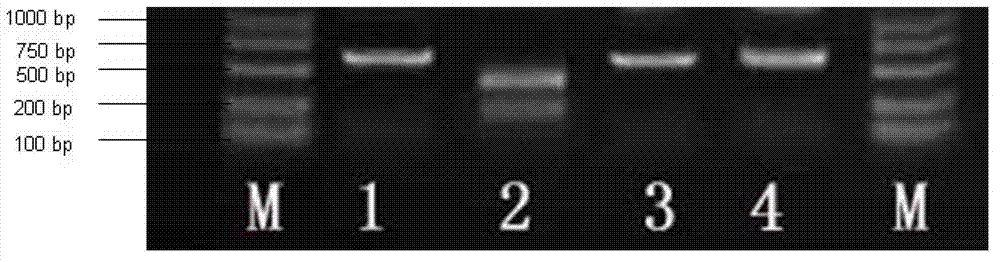
[0072] Amplify F with primers 2Population sample DNA. The reaction system includes 2 μl 10×PCR buffer (100 mM Tris-HCl pH 8.0, 15 mM MgCl2, 500 mM KCl, 1% TritonX-100), 0.2 mM dNTPs, 0.2 μM upstream and downstream primers, 50-100 ng sample DNA and 0.625 U Taq enzyme.
[0073] The reaction program was: pre-denaturation at 94°C for 5 min, cycle (94°C for 30 s, 56°C for 30 s, 72°C for 1 min) 35 times, and finally extension at 72°C for 10 min. The PCR product was digested with SpeI (TaKaRa), and the digestion reaction system was 20 μl, including: 17.5 μl PCR product, 2 μl 10× M buffer, 0.5 μl SpeI. Digested at 37°C f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com