Coronary heart disease susceptibility substantially-associated single nucleotide polymorphism (SNP) sites of susceptible region chr5p15, and applications thereof
A single nucleotide polymorphism and polymorphic site technology, applied in the determination/testing of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the lack of identification methods, little knowledge of the exact genetic molecular mechanism, etc. problem, to achieve the effect of reducing the incidence of
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
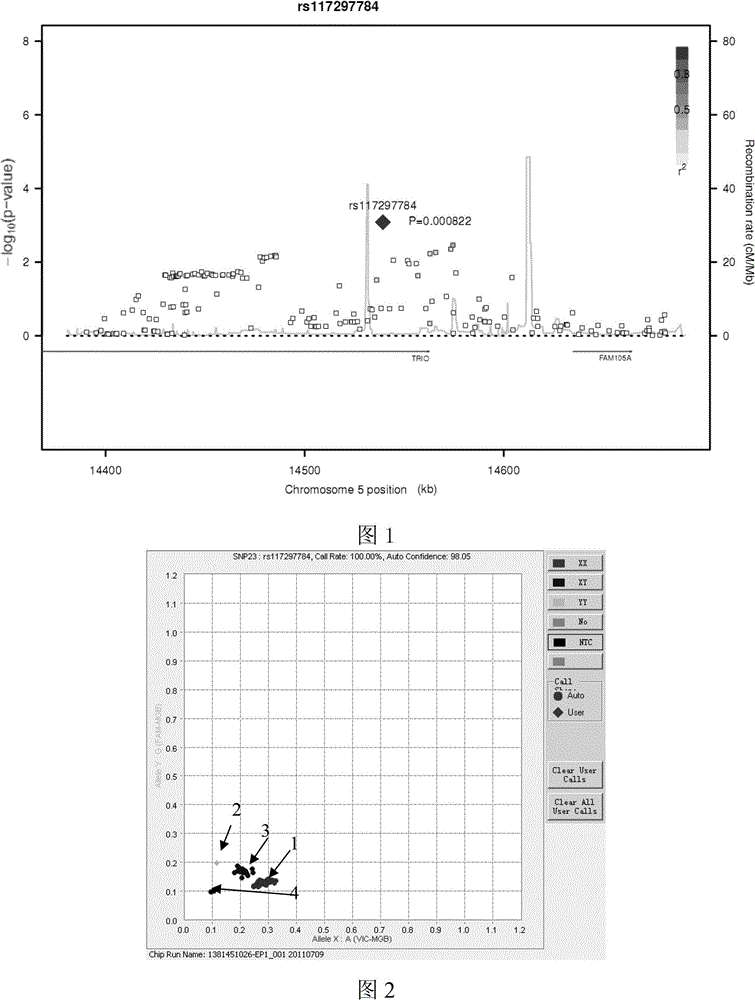
[0021] Example 1 Determination of single nucleotide polymorphism sites, alleles and risk alleles thereof that are significantly associated with susceptibility to coronary heart disease:
[0022] 1. Method
[0023] Firstly, 1515 patients with coronary heart disease and 5019 normal people were selected as the research objects in the Chinese Han population. Using Affymetrix's genome-wide chip (AffymetrixAxiom TM Genome-Wide CHB 1 ArrayPlate) was detected by the BIO-X Center of Shanghai Jiao Tong University. The genotype data of 1515 patients with coronary heart disease and 5019 normal people were obtained by whole genome microarray SNP. The significance level (P value) of the association between each site in the SNP chip and coronary heart disease was obtained through the statistical analysis of genome-wide association (GWAS), and the potential association with coronary heart disease was found according to the significance level (P value less than 0.001) Chromosomal regions, ...
Embodiment 2
[0036] Example 2 Determination of the most significant representative single nucleotide polymorphism loci associated with susceptibility to coronary heart disease
[0037] 1. Method
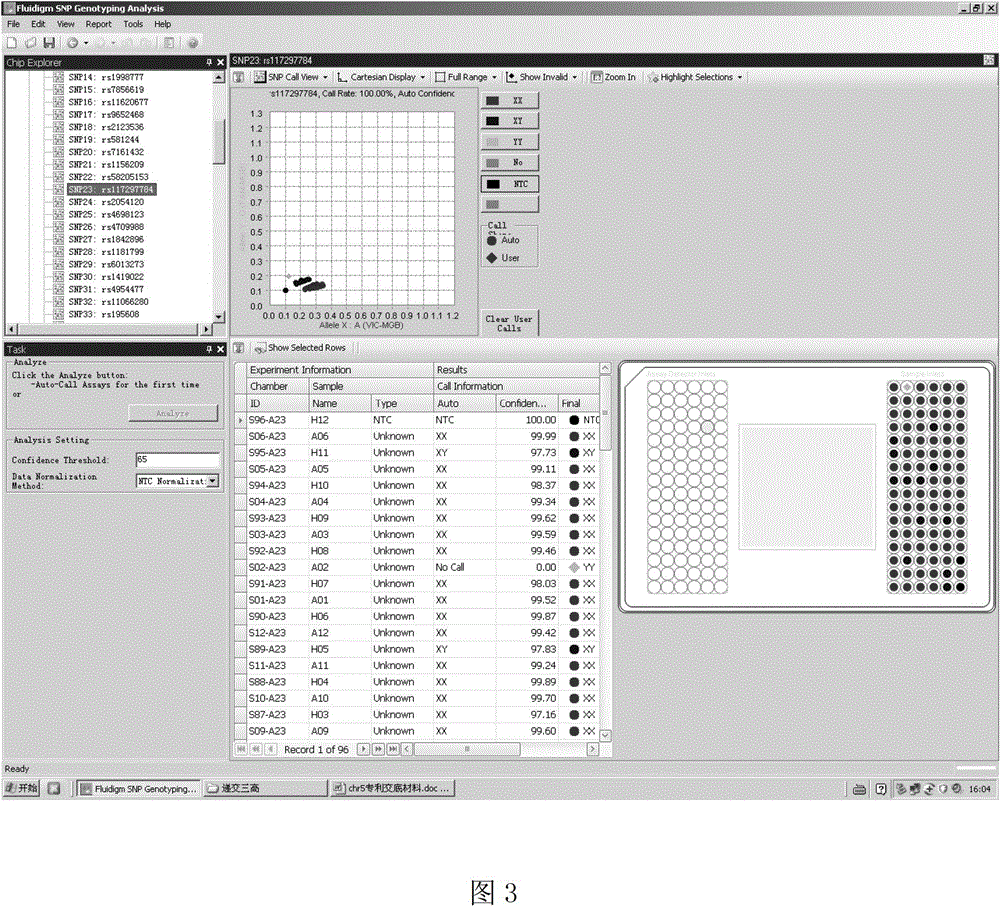
[0038] Use Haploview software to calculate the linkage disequilibrium (Linkage disequilibrium, LD) situation between the chromosomal region SNP sites that embodiment 1 gains, use r 2 Represents (0-1). Representative loci with strong linkage disequilibrium were selected and further validated in 11211 patients with coronary heart disease and 7286 control samples by using the FludigmEP 1 gene analysis system (FludigmEP 1 TM GENETIC ANALYSIS system), Taqman MGB probe method genotyped 11211 cases and 7286 control samples, combined the typing results of all 12726 cases and 12305 control samples, analyzed its association with coronary heart disease, and determined the correlation with coronary heart disease Representative SNP loci most significantly associated with susceptibility.
[0039] 2. Inclusi...
Embodiment 3
[0077] Example 3 Use of single nucleotide polymorphism sites significantly associated with susceptibility to coronary heart disease for assessing susceptibility to coronary heart disease
[0078] 1. Method
[0079] Three haplotypes were constructed from the 5p15 region rs117297784, rs1058312, rs9942367, rs17500919, and rs11133787. According to the genotype data of 1515 cases and 5019 control samples obtained by the whole genome chip, these SNP sites and a series of SNPs were analyzed The frequency difference of the haplotypes composed of the loci is different between patients and normal people, and the relationship between the variation of the specific bases at each locus and the haplotype differences composed of them and the susceptibility to coronary heart disease can be obtained. Also apply Fludigm EP1 TM GENETIC ANALYSIS system, Taqman MGB probe method conducts genotyping experiments on the population to be tested, which can determine the susceptible population of coronar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com