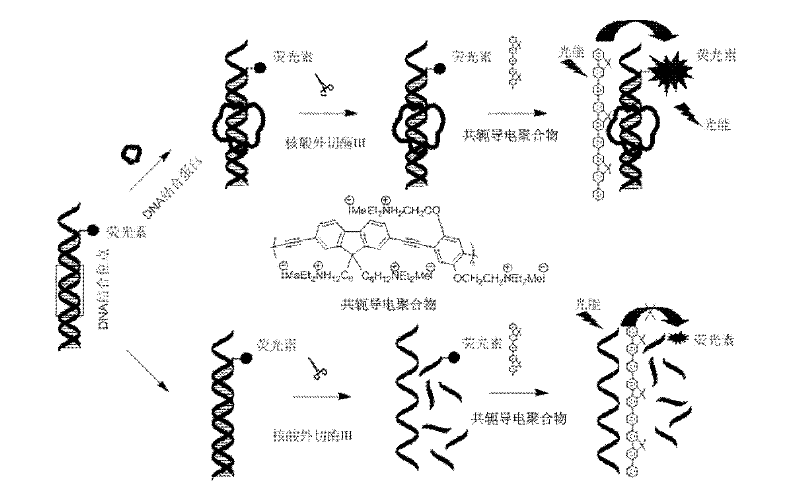
Fluorescence detection method for detecting desoxyribonucleic acid binding protein
A deoxyribonucleic acid and protein-binding technology, which is applied in the fields of fluorescence/phosphorescence, biological testing, and material inspection products, can solve the problems of complicated experiments, contamination of radioactive substances, and low analysis efficiency, and achieve high selectivity and sensitivity, shorten The effect of detection time and high analysis efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0040] a) Preparation of fluorescein-labeled nucleic acid probes
[0041] NF-κB nucleic acid probe:
[0042] A: 5′-AGTTGA GGGGACTTTCC CAGGCTTTTT-3′
[0043] B: 3′-TCAACTC CCCTGAAAGG GTCCG-FAM-5′
[0044] Where ______ is the binding site of NF-κB, and FAM is fluorescein.
[0045] When preparing NF-κB nucleic acid probes, mix equimolar solutions of two oligonucleotide chains A and B in hybridization buffer (10mM Tris-HCl, 100mM NaCl, pH 8.0), incubate at 95°C for 10 minutes, and cool slowly to room temperature. Then the probe solution was diluted with hybridization solution to a concentration of 10 μM, and stored at 4° C. for future use.
[0046] b) The labeled nucleic acid probe binds to nuclear factor NF-κB
[0047] Add nuclear factor NF-κB protein to deoxyribose nucleic acid binding buffer, incubate at 37°C for 10 minutes, then add 10 μM nucleic acid probe, and incubate at 37°C for 20-30 minutes.
[0048] c) Exonuclease III digestion
[0049] The 10× digestion reac...
Embodiment 1
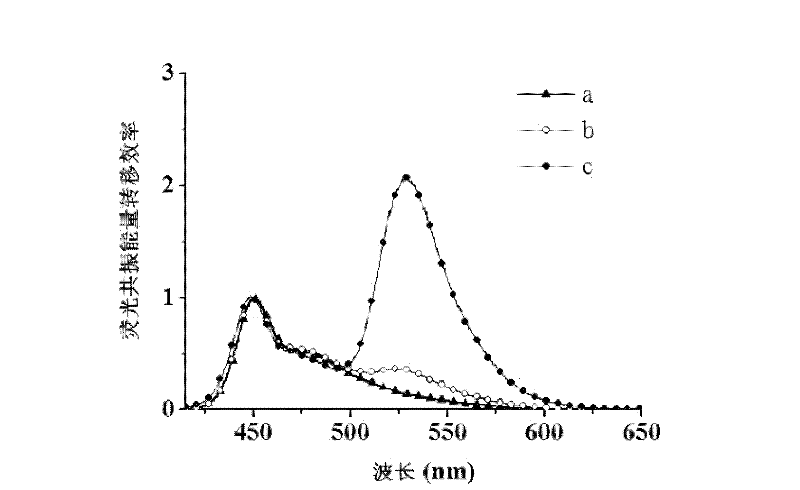
[0052] Example 1 Detection of nuclear factor NF-κB protein.
[0053] Detection 1: Add nuclear factor NF-κB protein to deoxyribonucleic acid binding buffer, incubate at 37°C for 10 minutes, then add 10 μM double-stranded nucleic acid probe, and incubate at 37°C for 20-30 minutes. Add 10× enzyme digestion reaction solution and 20U / μL exonuclease III to the reaction system, and incubate at 37°C for 5-10 minutes. After the enzyme cleavage reaction, add 0.4M ethylenediaminetetraacetic acid solution to terminate the enzyme cleavage reaction of exonuclease III, and then add it to the detection buffer containing water-soluble conjugated polymer, the detection excitation wavelength is 404nm, and the emission The wavelength is 410-650nm. Since the labeled nucleic acid probe will undergo a sequence-specific binding reaction with the nuclear factor protein NF-κB, the formed "nucleic acid probe / deoxyribonucleic acid binding protein" complex provides space protection for the nucleic acid p...
Embodiment 2
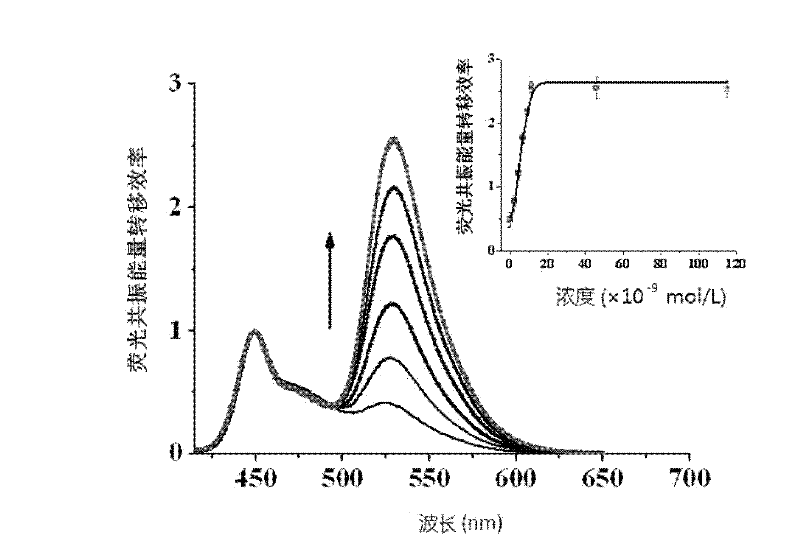
[0060] Example 2 detects different concentrations of nuclear factor protein NF-κB.
[0061] Different concentrations of nuclear factor NF-κB protein were detected, and other conditions were the same as detection 1 in Example 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com