Method for rapid detection of enterobacter sakazakii
An Enterobacter sakazakii, fast technology, applied in measuring devices, instruments, analysis by nuclear magnetic resonance, etc., can solve the problems of high false positive and false negative rates in PCR reactions, and achieve good reliability, simple processing, and low requirements. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Preparation of Immunized (Biologically Functionalized) Superparamagnetic Nano Magnetic Beads
[0031] 1. Ferric oxide (Fe 3 o 4 ) synthesis and encapsulation and amino functionalization
[0032] ①, 20mM (3.25g) of ferric chloride, 120mM (12.3mL) of acetylacetone, and 20mL of distilled water were mixed and stirred for 15 minutes, then 6mL of triethylamine was added and filtered to obtain a red powder, which was dissolved in 30mL of absolute ethanol and distilled water ( V:V 7:3), heated to 80°C, cooled to room temperature and recrystallized to obtain iron acetylacetonate.
[0033] Add 1.06g of ferric acetylacetonate into the system of 15mL of phenyl ether and 15mL of oleylamine, stir it with a magnetic force, and continuously raise the temperature to 115°C (the temperature range can be 110-120°C), keep it for 30 minutes, vacuumize, and continue to heat up to 180°C (Temperature range can be at 175~185°C), keep for 30 minutes, continue to heat up to 300°C (tem...
Embodiment 2
[0038] The immunized superparamagnetic nano-magnetic beads prepared in Example 1 were dispersed in phosphate buffer (0.01 mol / L, pH=7.2) at a concentration of 0.8 mg / mL (experimentally optimized data).
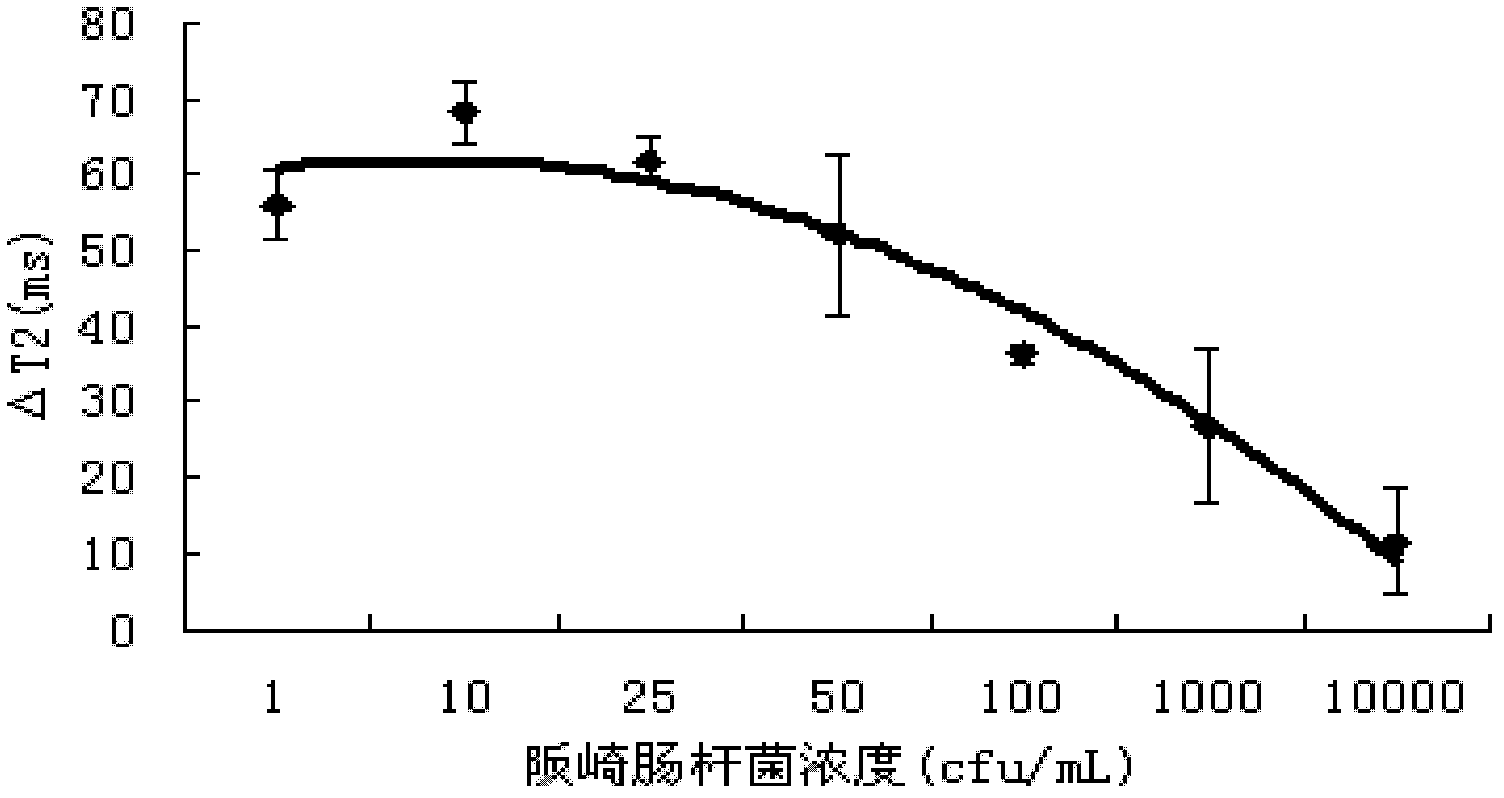
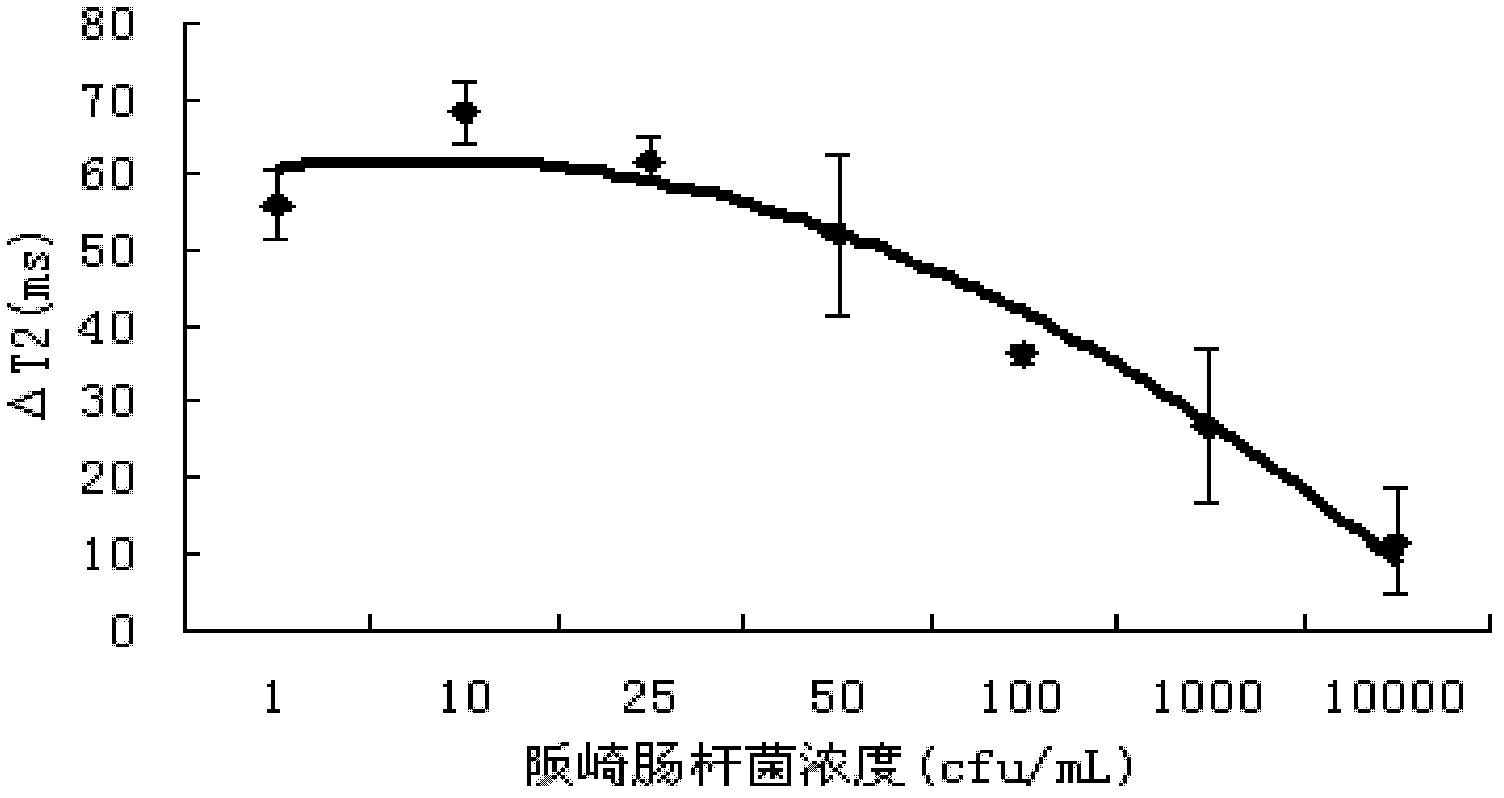
[0039] Add 225uL of the above-mentioned magnetic beads to 75uL of E. sakazakii containing different concentrations (1cfu / ml, 10cfu / ml, 25cfu / ml, 50cfu / ml, 10 2 cfu / ml, 10 3 cfu / ml, 10 4 cfu / ml) dairy product samples, add 2.7 mL of stabilizer 2% milk, and incubate at 37° C. for 45 min to 60 min (data optimized in the experiment). A non-bacterial dairy product sample was used as a control.
[0040] Add the processed sample into the NMR tube with a diameter of 3cm, put the NMR tube into the NMR analyzer, and adjust the parameters, D1(us): 100; D2(us): 200; D0(ms): 100; TD : 20000; SW (kHz): 75.0; DFW (kHz): 30.0; RG: 2; Ns: 8; C1: 2000; (T 2 ), calculate ΔT 2 value. The result is as figure 1 shown.
Embodiment 3
[0042] The immunized superparamagnetic nano-magnetic beads prepared in Example 1 were dispersed in phosphate buffer (0.01 mol / L, pH=7.2) at a concentration of 0.8 mg / mL (experimentally optimized data).
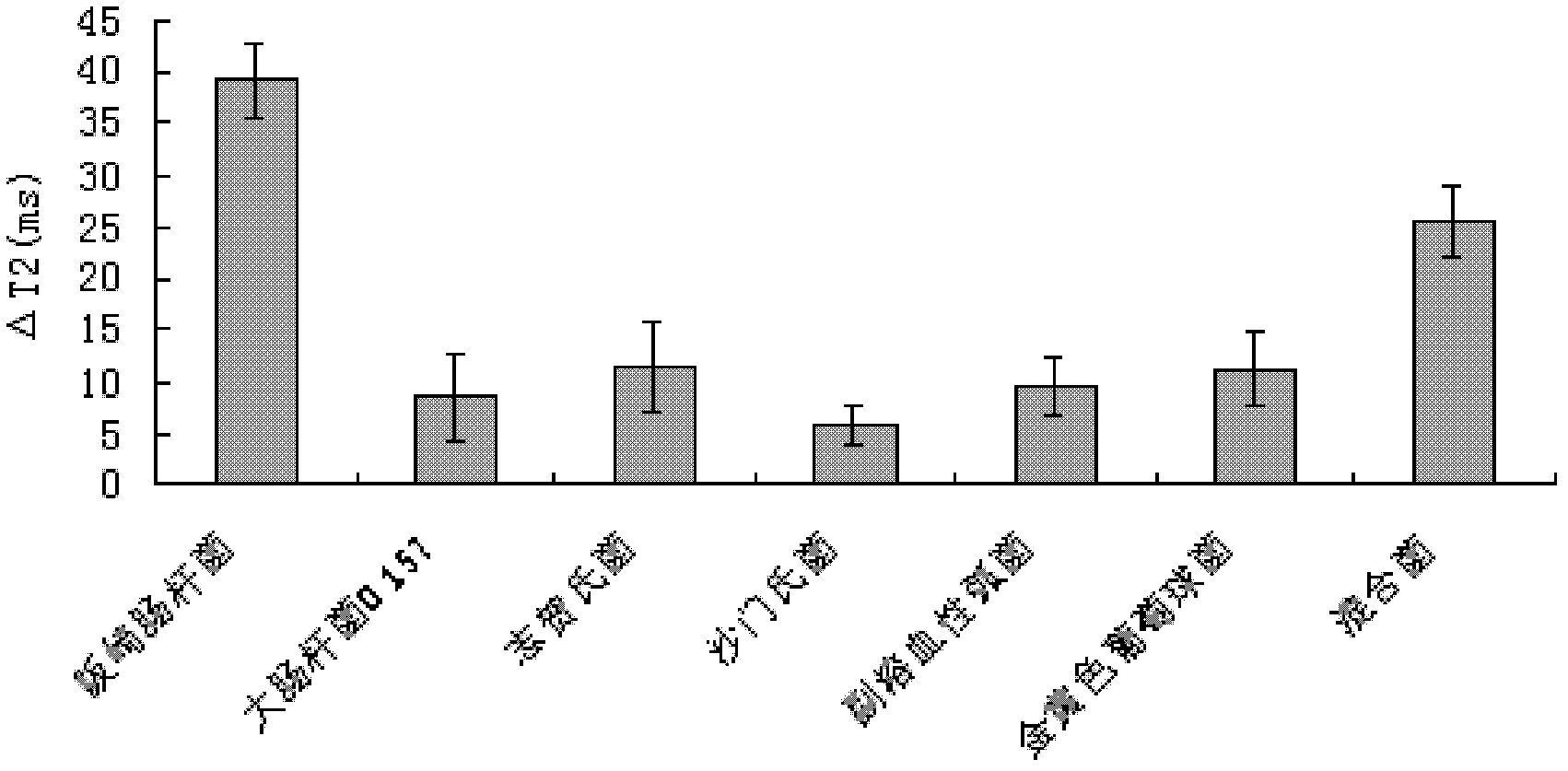
[0043] Take 225uL of the above-mentioned magnetic beads and add them to 75uL of dairy product samples that have been incubated for 2h (Enterobacter sakazakii concentration 10 2 cfu / ml), add 2.7 mL of stabilizer 2% milk, and incubate at 37° C. for 45 min to 60 min (data optimized in the experiment). A non-bacterial dairy product sample was used as a control.
[0044] Add the processed sample into the NMR tube with a diameter of 3cm, put the NMR tube into the NMR analyzer, and adjust the parameters, D1(us): 100; D2(us): 200; D0(ms): 100; TD : 20000; SW (kHz): 75.0; DFW (kHz): 30.0; RG: 2; Ns: 8; C1: 2000; (T 2 ), calculate ΔT 2 value.
[0045] And use the same conditions to detect the same concentration (10 2 cfu / ml) of Escherichia coli O157, Shigella, Salmonella, Vibrio p...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com