Methods for the detection of colorectal tumors
A colorectal and tumor technology, applied in the field of colorectal tumor determination, can solve problems such as non-quantitative detection, and achieve the effect of simple detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] (DNA Extraction from Surgical Resection Frozen Specimens)
[0072] DNA was extracted from surgical resection frozen specimens using All Prep DNA / RNA Mini Kit (Qiagen, Hilden, GERM) from 252 samples of normal colorectal mucosa and 320 samples of colorectal cancer. Add 600 μl of RLT plus buffer solution attached to the product to a 2ml tube, add 6 μl of β-mercaptoethanol, add a surgical excision frozen specimen, add 1 stainless steel bead (Stainless Steel Beads) 5 mm (manufactured by Qiagen), and use Qiagen Mixer Mill MM300 (manufactured by Qiagen) ultrasonically disrupted the specimen at 30 Hz for 10 minutes. The homogenized cell lysate was added to an All Prep DNA spin column (manufactured by Qiagen), and centrifuged at 10,000 rpm for 30 seconds. Place a new All Prep DNA spin column on a 2ml collection tube and incubate at room temperature or 4°C.
[0073] Add 500 μl of AW1 buffer (included with the product) to the incubated DNA spin column, centrifuge at 10,000 rpm f...
Embodiment 2
[0096] (Using TWIST1 gene methylation as an indicator to detect colorectal adenoma)
[0097] Using the same protocol as in Example 1, it was verified whether methylation of the TWIST1 gene could be a marker for colorectal adenoma, which is a benign tumor arising in the large intestine.
[0098] Of the 189 samples that were paraffin-embedded specimens of formalin-fixed colorectal adenomas, only the tumor portion was recovered by microdissection, and DNA was extracted using a Qiagen DNA FFPE Tissue kit (manufactured by Qiagen). The extraction method was performed according to the protocol accompanying the product.
[0099] (Preparation of samples)
[0100] The control samples were prepared according to Example 1. The positive control of the methylation array was prepared from placenta-derived DNA (Sigma) treated with SssI methyltransferase (New England Biolabs), respectively, and extracted from peripheral blood lymphocytes. DNA was prepared as a positive control for unmethylat...
Embodiment 3
[0119] (Use of colorectal cancer cell line)
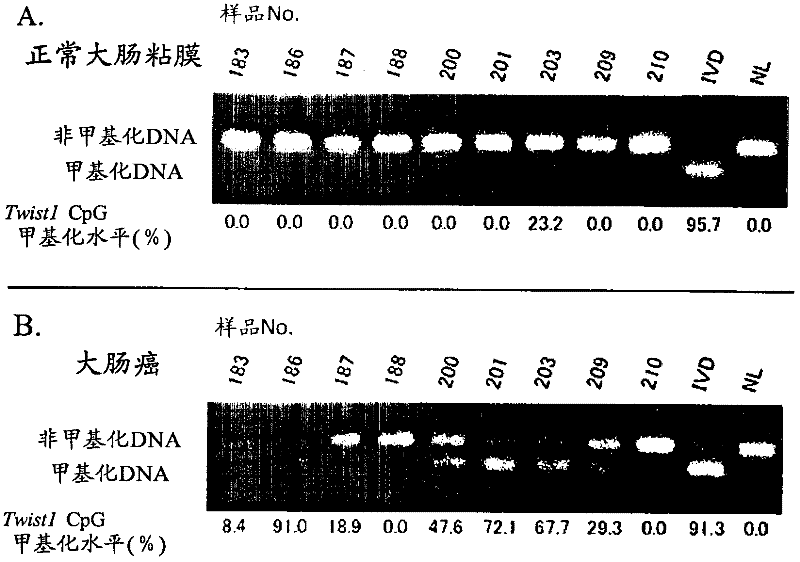
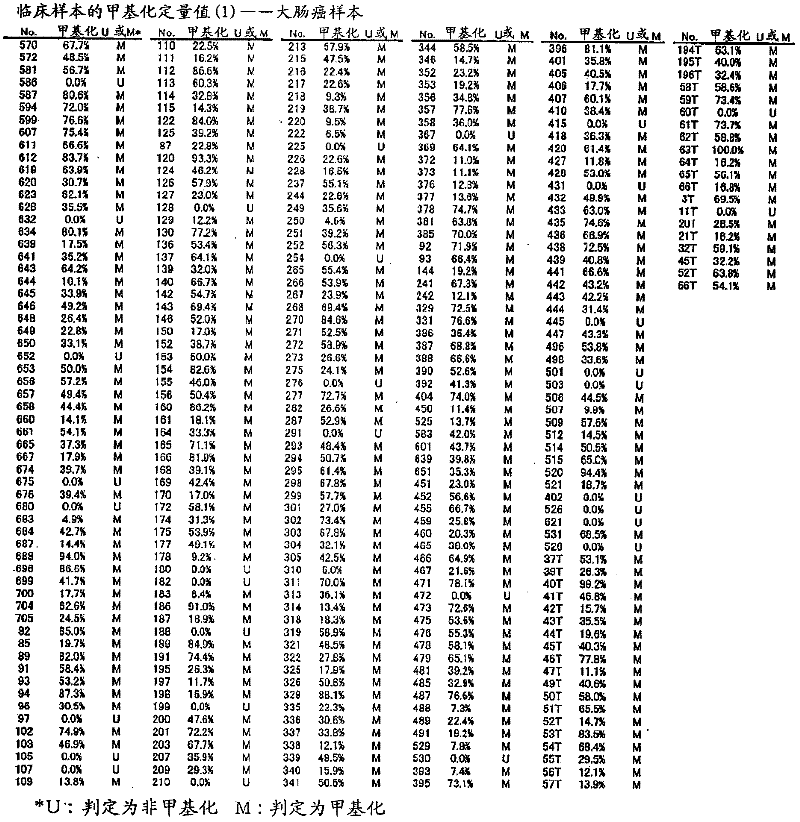
[0120] For the CpG sequence in the -477 to -747 region of the TWIST1 gene using a colorectal cancer cell line, specifically, CGCG at positions 57 to 60 in the nucleotide sequence shown in SEQ ID NO: 1 and CpG at positions other than that methylation was studied. As colorectal cancer cell lines, DLD-1, HT-29, HCT-116, and RKO were used, NL derived from peripheral blood lymphocytes was used as a negative control for methylation, and NL derived from peripheral blood lymphocytes was used as a positive control for methylation. IVD of methyltransferase-treated DNA. For colorectal cancer cell lines (DLD-1, HT-29, HCT-116, RKO), the methylation profiles of 10 clones (the methylation profiles of each of the 10 cells) were studied respectively. The methylation profiles of 5 clones were studied separately for the positive (IVD) and negative controls (NL). Figure 24 It is a graph showing the distribution of CpG methylation in TWIST1 gene -...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com