Sextuple PCR (polymerase chain reaction) detection method of portunus trituberculatus miers microsatellite marker
A technology of Portunus trituratus and microsatellite markers, which is applied in the direction of determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve problems such as time-consuming, complicated operation, etc., and achieve the effects of improving efficiency, shortening experimental time, and being easy to detect.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] A six-fold PCR construction method of Portunus trituberculatus microsatellite markers of the present invention is as follows:
[0033] 1. Based on the microsatellite core sequence in the Portunus trituberculatus microsatellite enrichment library provided by the present invention, use Primer Premier 6.0 software to design corresponding PCR primers at the two ends of its microsatellite core sequence to obtain a series of PCR primers with PCR amplification. Amplified SSR markers of varying product sizes.
[0034] 2. Use Primer Premier 6.0 software to evaluate between primers, and select the primers that show most of the ΔG (Gibbs of energy change) value above -4 for use, and the ΔG (Gibbs of energy change) value above -4 is mainly explained In the multiplex PCR system, it is difficult for a single pair of primers to form primer dimers and have a low hairpin structure, so as not to affect the efficiency of the PCR reaction. Through the matching and combination of 22 pairs ...
Embodiment 2
[0049] This embodiment is aimed at Mg 2+ Concentration optimization experiment was carried out, the experimental process is as follows:
[0050] Steps (1) and (2) are the same as in Example 1.
[0051] The PCR reaction system in step (3) is: 10×Buffer 1.5 μ L; 2.5 mmol / LdNTP 1.5 μ L; Ptr33a of the present invention, Ptr95, Pot42, Pot44 and Pot57 primers are respectively 0.4 μ L, Pot34 positive and negative primers respectively 0.5 μ L, each Primer concentration is 10pmol / μL; 50ng / μL Portunus trituberculatus genomic DNA 0.6μL; 5U / μL Taq polymerase 0.12μL; ddH 2 O supplemented to 15 μL; 15 mmol / L MgCl 2 The volume was optimized with a gradient of 0.8, 1.0, 1.2, 1.4 μL.
[0052] The reaction program used the conventional PCR reaction program: pre-denaturation at 94°C for 5 min, denaturation at 94°C for 40 s, annealing at 60°C for 1 min, extension at 72°C for 1 min, and a total of 25 cycles. Finally, extend at 72°C for 5 minutes, store at 4°C and end the program.
[0053] The...
Embodiment 3
[0055] This embodiment selects the above-mentioned optimized reaction program and Mg 2+ Volume, the six-fold PCR detection method concrete steps of a kind of Portunus trituberculatus microsatellite marker of the present invention are as follows:
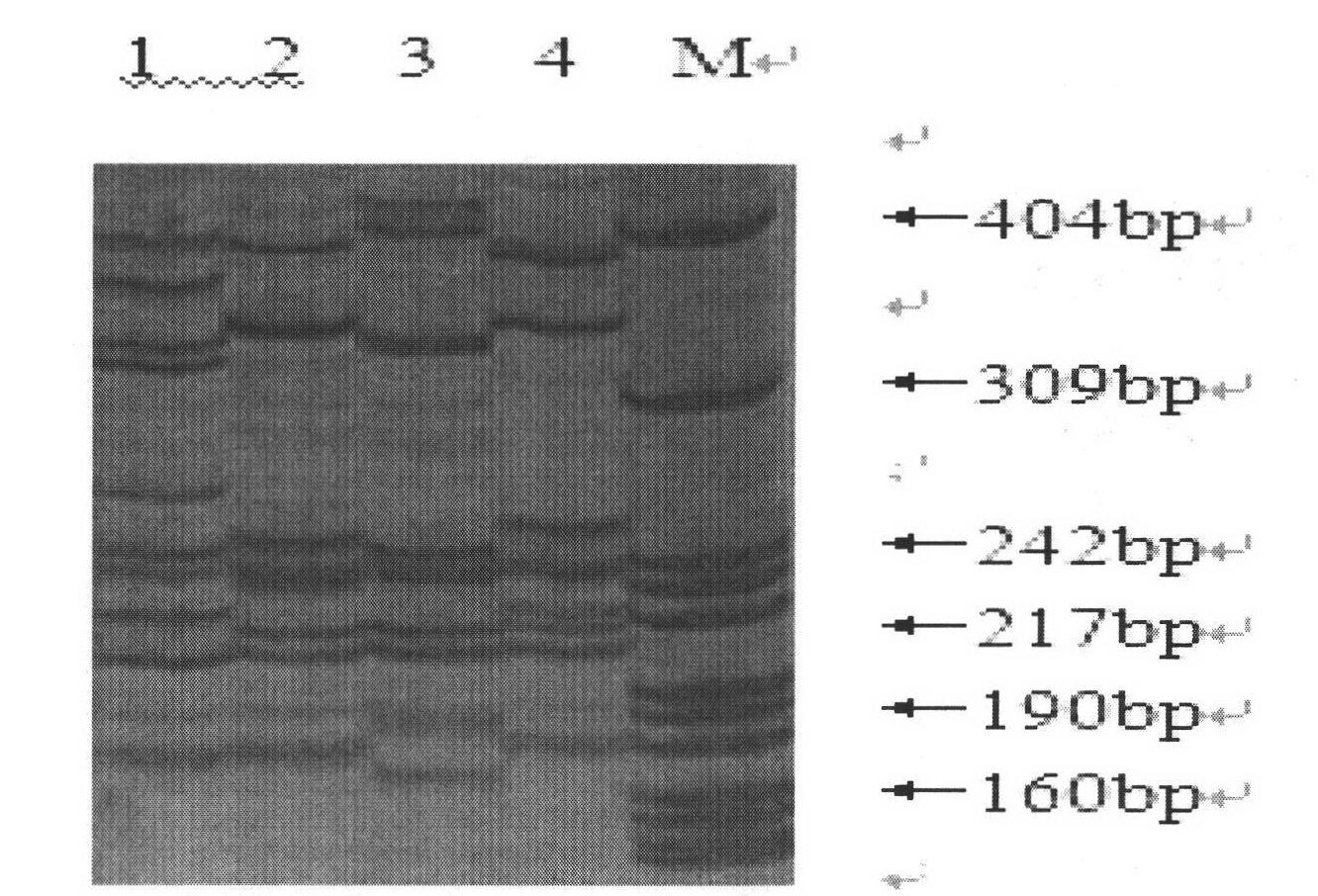
[0056] (1) The selected six primer sequences of the present invention are shown in Table 2 below:
[0057] Table 2: Six primer sequences and their annealing temperatures and accession numbers of their core sequences
[0058]
[0059] (2) Genomic DNA extraction of Portunus trituberculatus: the large claw muscle tissue of Portunus trituberculatus was taken, and the genomic DNA of Portunus trituberculatus was extracted by the method of phenol / chloroform. The integrity of genomic DNA was detected by 1% agarose gel electrophoresis, the concentration of genomic DNA was detected by RNA / DNA spectrophotometer and diluted to a final concentration of 50ng / μl, and stored at -20°C for later use.
[0060] (3), PCR reaction system: 10×Buffer 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com