Nosema bombycis SYBT Green fluorescence quantitative PCR (Polymerase Chain Reaction) detection method as well as kit and specific primer thereof
A microsporidia, fluorescence quantitative technology, applied in the direction of microbial determination/inspection, fluorescence/phosphorescence, biochemical equipment and methods, to avoid missed detection and false detection, enhanced sensitivity and specificity, good repeatability and stability sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
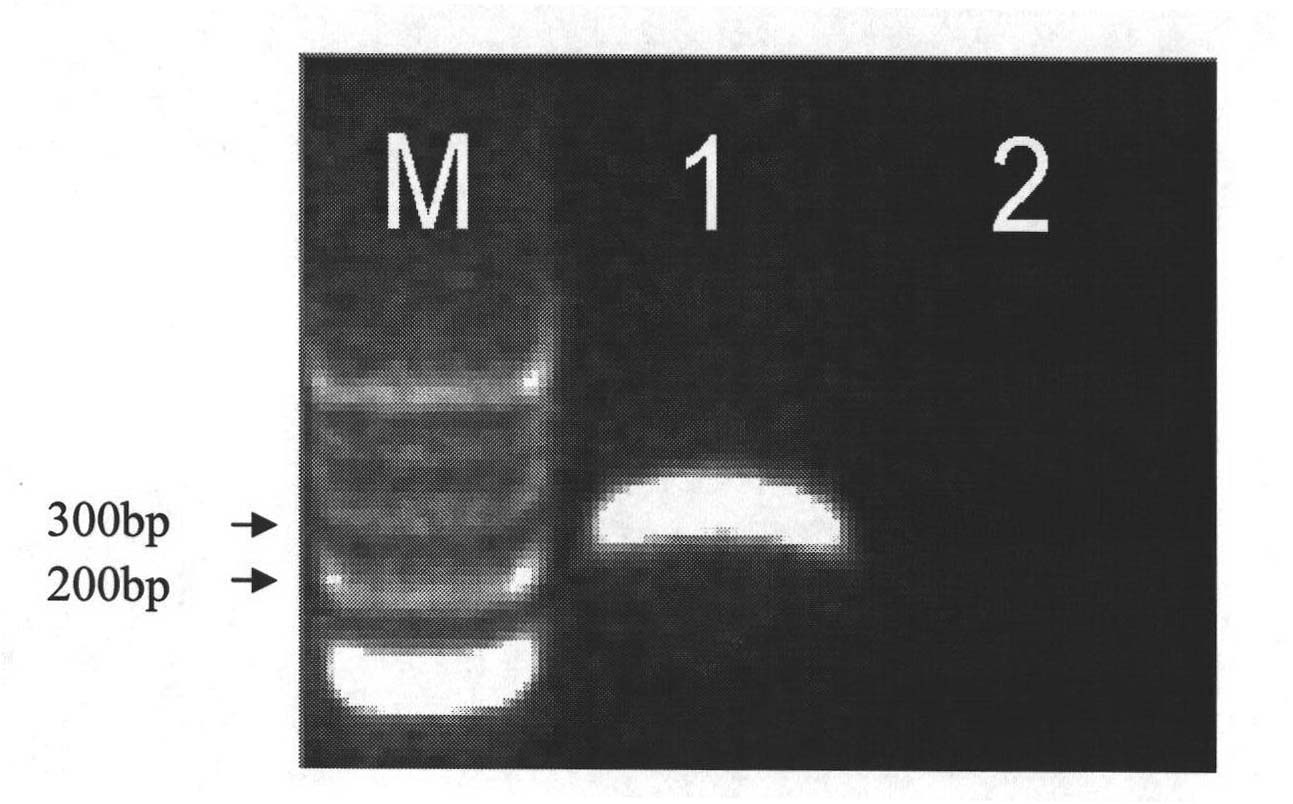
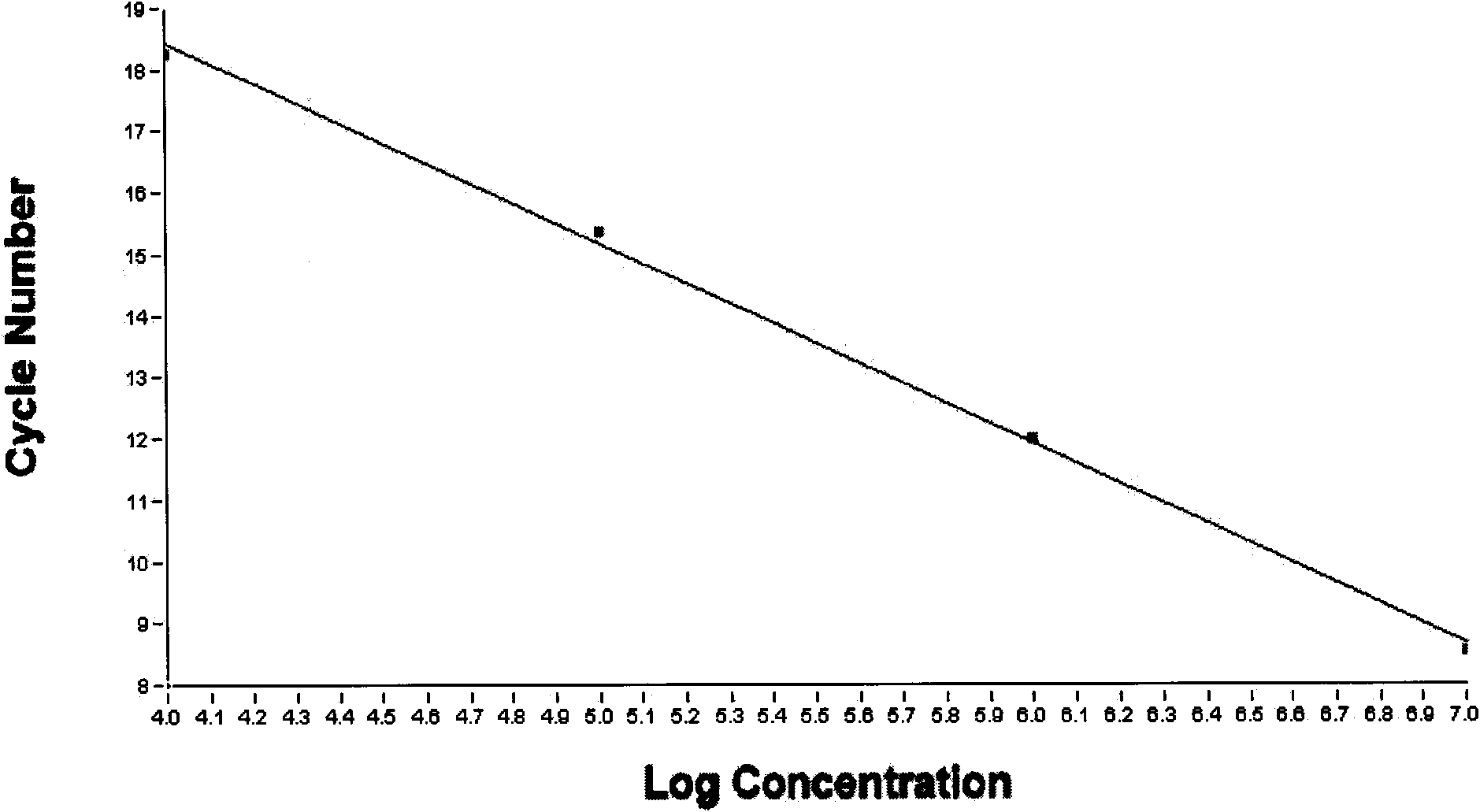
Image
Examples
Embodiment 1
[0033] Example 1: Specimen collection
[0034] Applicable specimen types include samples such as silkworms, silkworm eggs and feces. Take an appropriate amount of suspected disease materials such as silkworms, silkworm eggs or feces from the scene, seal them, label them, and send them for inspection as soon as possible or in a short-term refrigerated storage.
Embodiment 2
[0035] Embodiment 2: the extraction of silk microsporidia DNA
[0036] Take the above specimen and add appropriate amount of normal saline to grind, filter the homogenate with gauze, collect the filtrate, and use differential centrifugation (500rpm, centrifuge for 2 minutes, discard the precipitate, then centrifuge the supernatant at 5000rpm for 10 minutes), discard the supernatant, and divide Centrifuge quickly until the precipitate is off-white, suspend it with an appropriate amount of normal saline, and obtain a crude extract of Nb spores. Take 0.5ml (10 8 individual / ml) Microsporidia suspension, centrifuge at 5000rpm for 5 minutes, remove the supernatant, keep the spores, add 0.2mol / L KOH 0.25ml at the same time, mix thoroughly, induce at 27℃ for 1 hour, centrifuge at 5000rpm for 5 minutes, and remove clear. Resuspend with 0.5ml of normal saline and place at 25°C for 30 minutes. Centrifuge at 5000rpm for 5 minutes, add 50μl of nucleic acid extraction solution to the pre...
Embodiment 3
[0037] Embodiment 3: the amplification of silk microsporidia DNA
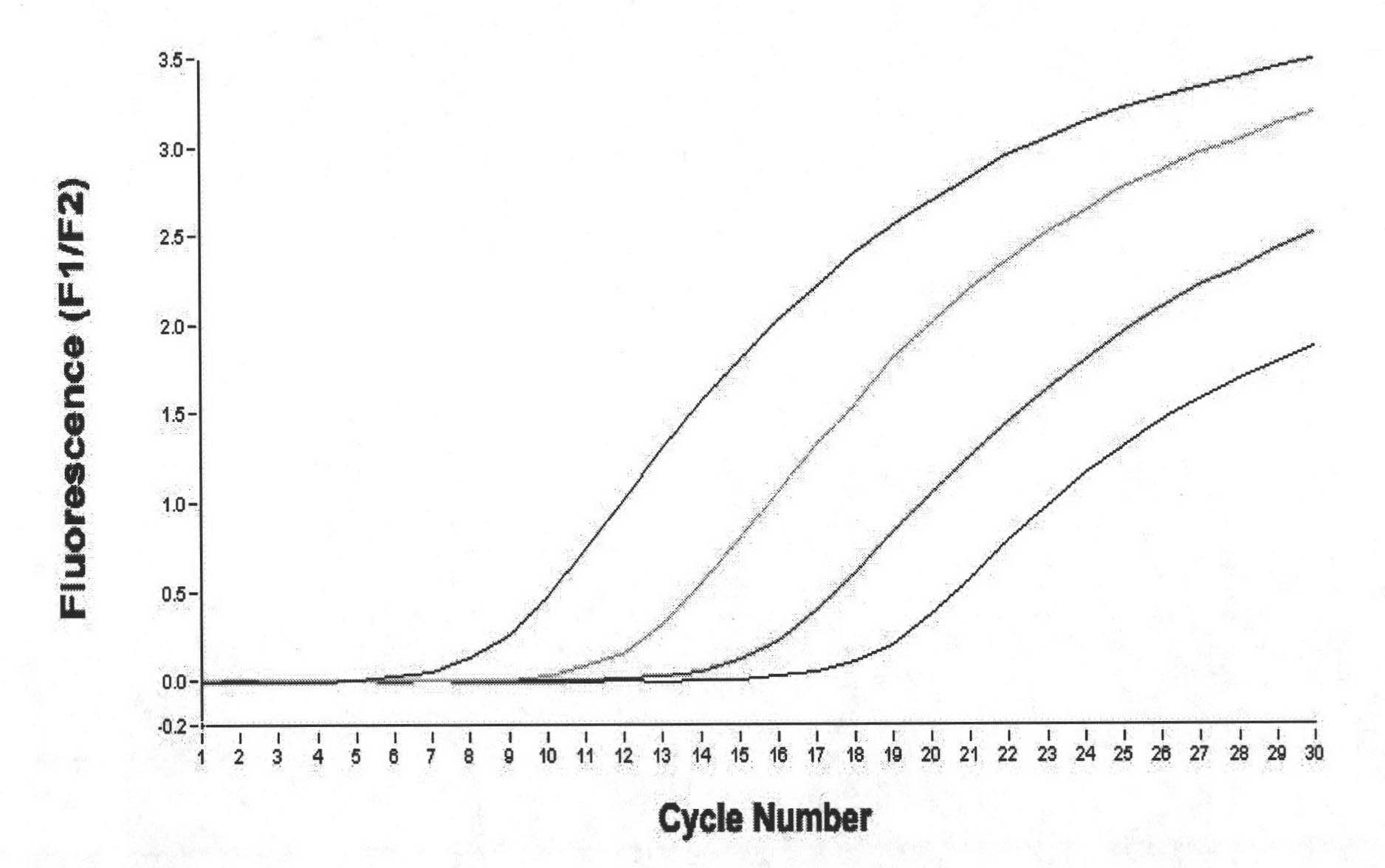
[0038] Take out the Nb-PCR MIX from the kit, melt at room temperature and shake to mix, then centrifuge at 10,000rpm for 10 seconds. Each Nb-PCR MIX detection unit consists of 10 μl 2×SYBR Premix Ex Taq, 0.4 μl ROX ReferenceDyeII, 0.4 μl upstream primer (10 μM), 0.4 μl downstream primer (10 μM), 6.8 μl ddH 2 O composition. The sequence of the upstream primer is as follows: 5'-GGAAAGAAGAAAGGCCCAAG-3', and the sequence of the downstream primer is as follows: 5'-ATCTTCCAGCGTCGTTGATT-3'. 2×SYBR Premix Ex Taq and ROX Reference Dye II in the reaction system used to detect microsporidia above were provided by TaKaRa Bioengineering (Dalian) Co., Ltd.
[0039] Each test reaction system was prepared as follows: 18 μl of Nb-PCR MIX was placed in a clean 0.2ml PCR tube, and 2 μl of processed sample (extracted DNA) or positive standard or negative standard was added to the set PCR reaction tube, 10,000 Centrifuge briefly...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com