Method for identifying whether wheat to be tested is Pina-D1b deficient wheat and applications thereof
A deletion-type, wheat-based technology, applied in biochemical equipment and methods, recombinant DNA technology, and microbial assay/inspection, etc., can solve problems such as unqualified DNA quality or human error, and achieve faster screening efficiency and accuracy. The effect of increased reliability and credibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment 1, the mensuration of grain hardness of different wheat varieties
[0033]Detect the grain hardness of 12 wheat varieties (Durum Langdon, China Spring, Yunong 202, Shan 253, Gaocheng 8901, CMH82A.1294 / 2*KAUZ / / MUNIA / CHT O / 3 / MILAN, Mo 299, Mo 293, ink 215, ink 191, ink 181, ink 272, RDWG / MILAN). Grain hardness measurement method: measure the hardness value of 300 grains of wheat samples with a single-grain grain hardness instrument (SKC84100, Perten, Sweden), determine the hardness level of the kernels according to the measurement results, and record the thousand-grain weight and kernel diameter at the same time. The experiment was repeated three times, and the results were averaged.
[0034] The results of SKCS analysis showed that among the 12 wheat varieties, China Spring belongs to soft wheat, and the remaining 11 wheat varieties are all hard wheat (hardness above 60). The average hardness values of SKCS are: China Spring is 12, Yunong 202 is 60, Shaanx...
Embodiment 2
[0035] Embodiment 2, the discovery of specific primer pair
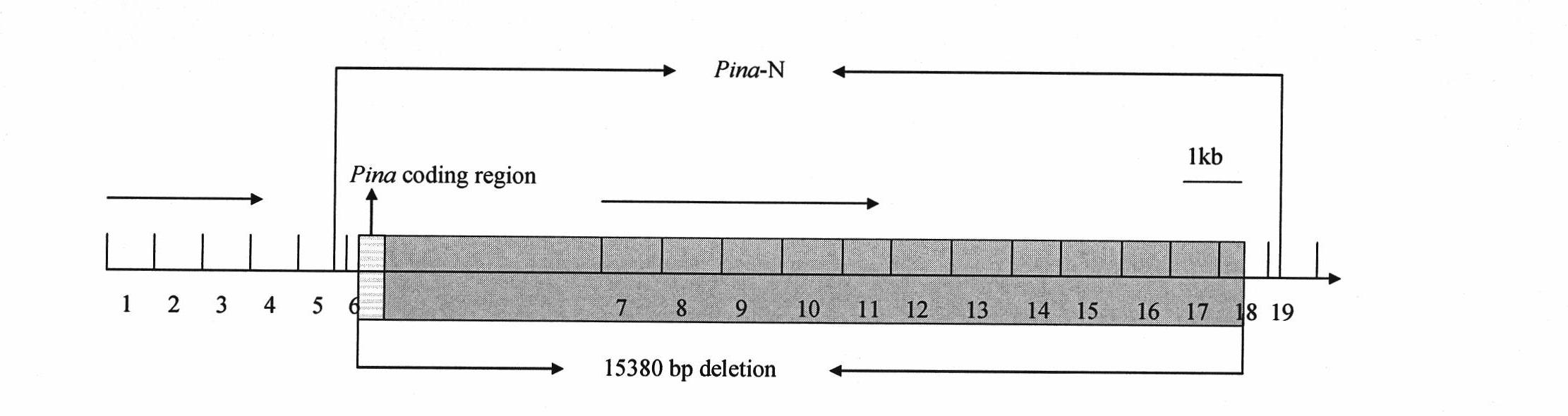
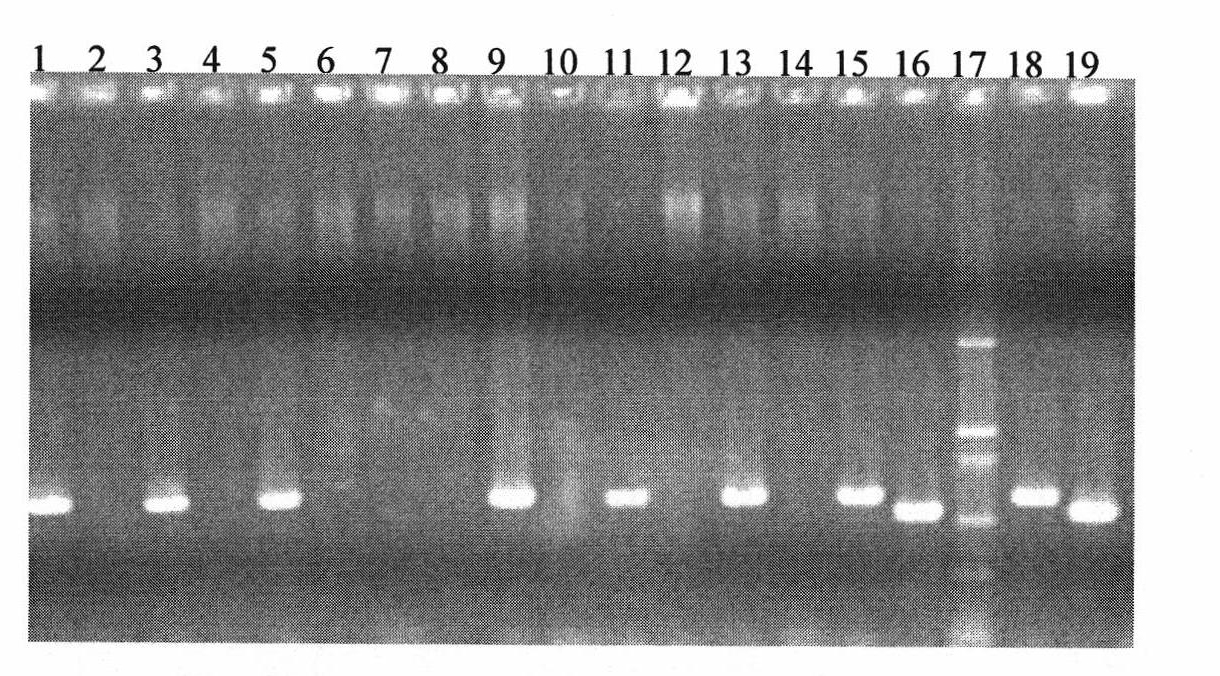
[0036] Taking Shan 253 and Gaocheng 8901 as examples, the target fragments were amplified by the primer walking method. Primer design as figure 1 shown. Among them, the 1st, 2nd, 3rd, 4th, 5th and 19th pair of primers can amplify the target fragment, and the 6th, 7th, 8th, 9th, 10th, 11th, 12th, 13th, 14th, 15th, 16th, 17th and 18th pair The primers cannot amplify the target fragment, indicating that the missing fragment occurs between the downstream of the fifth pair of primers and the upstream sequence of the 19th pair of primers, and then redesign specific primer pairs between the fifth pair of primers and the 19th pair of primers Amplification was performed to obtain a 776bp fragment. After the fragment was sequenced, compared with the same sequence of the wild-type Ha site (NCBI number: CT009735), it was found that the Pina-D1b type Ha site sequence lacked 15380bp, and the missing fragment was from the +23rd ...
Embodiment 3
[0040] Embodiment 3, application-specific primer pair identification Pina-D1b deletion type wheat
[0041] 12 wheat varieties: China Spring (soft wheat), Yunong 202 (hard wheat), Shan 253 (hard wheat), Gaocheng 8901 (hard wheat), Mo 299 (hard wheat), Mo 293 (hard wheat), Mo 215 (hard wheat), ink 191 (hard wheat), ink 181 (hard wheat), ink 272 (hard wheat), RDWG / MILAN, CMH82A.1294 / 2*KAUZ / / MUNIA / CHTO / 3 / MILAN (hard wheat ).
[0042] The 12 wheat varieties were identified as follows:
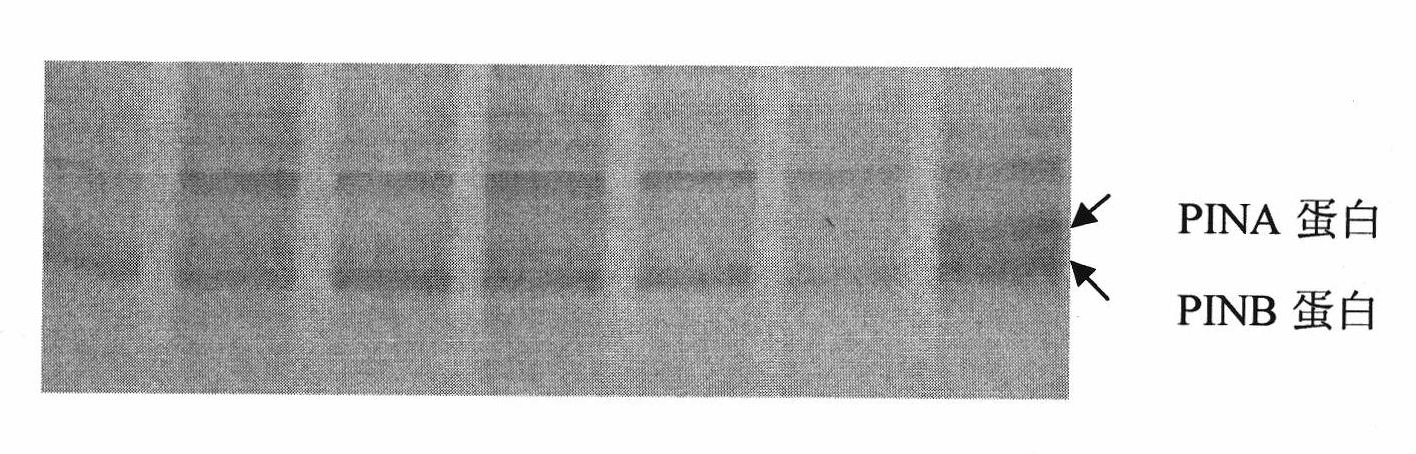
[0043] 1. SDS-PAGE to identify whether each wheat variety belongs to the type of PINA protein deficiency
[0044] 1. Extract grain protein
[0045] Take one seed of each variety of wheat, and extract the grain protein according to the following steps: 1) put it into a 2mL centrifuge tube after grinding, add 1mL pre-cooled TBS solution and 0.15mL 12% Triton-X114 (Sigma company) After mixing, place it at 4°C for 12-24h; 2) Centrifuge at 12000rpm for 3 minutes, transfer the supernatant to a 1.5mL c...
PUM
| Property | Measurement | Unit |
|---|---|---|
| hardness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com