Beads for high-throughput nucleic acid analysis
A technology of beads and functional groups, applied in the field of nucleic acid analysis, can solve problems such as difficult sample processing and side reactions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0163] The preparation of the beads has been disclosed in detail above.
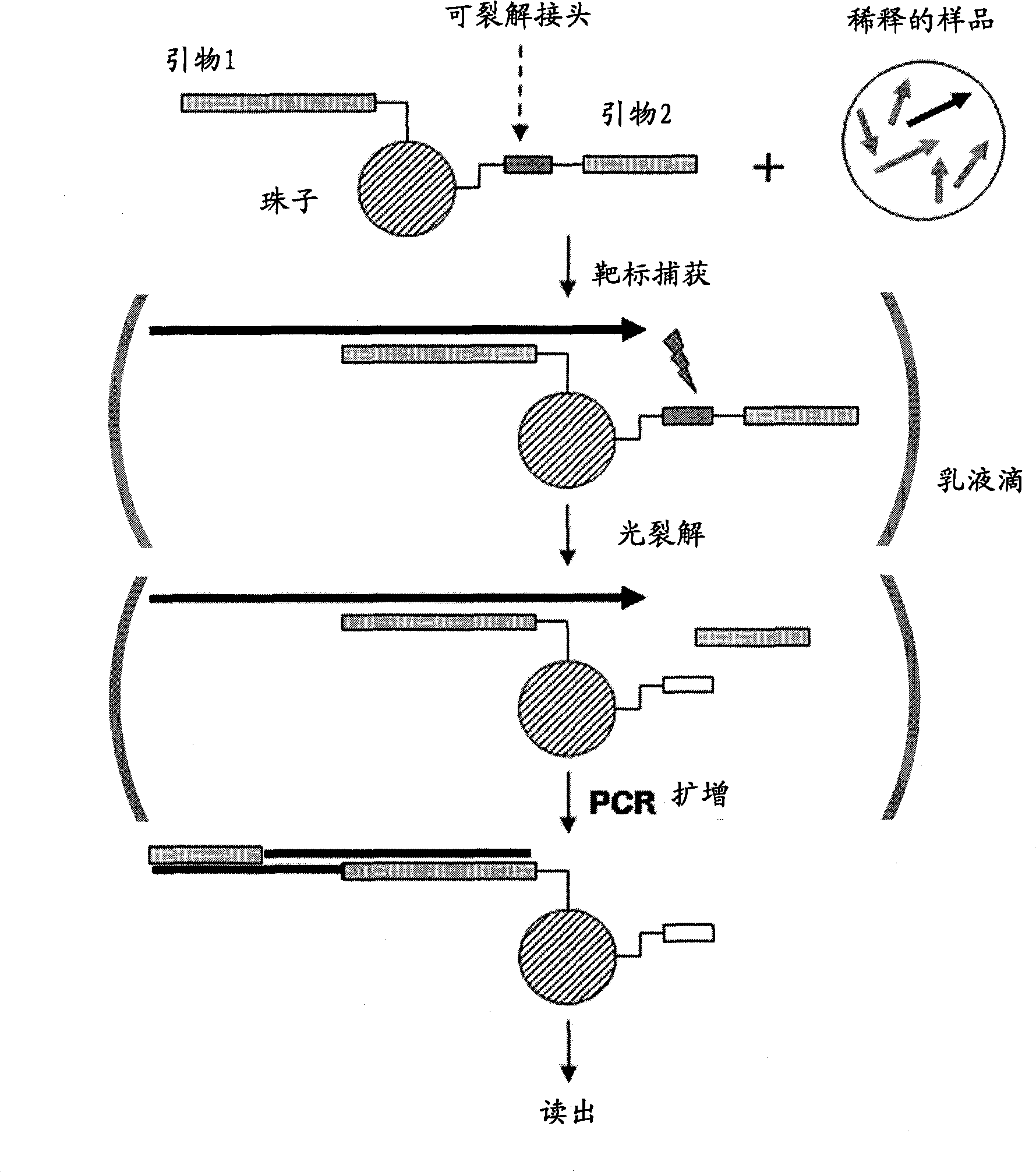
[0164] b) capture nucleic acid molecules of interest from the sample
[0165] The target molecules are then hybridized to beads containing cleavable and non-cleavable primers. Appropriate hybridization conditions in terms of appropriate buffer system and appropriate hybridization temperature are well known in the art, and can be optimized according to specific conditions (such as the length and sequence of specific amplification primers used). Preferably, hybridization is performed using a molar excess of beads compared to the sequence or sequences to be amplified in order to capture as much target nucleic acid as possible. In particular, a molar excess of 1:5 to 1:100, preferably 1:10 to 1:50, has proven to be particularly advantageous. If multiple different target sequences need to be detected and / or analyzed, it is necessary to use a bead library with corresponding multiple different primer pairs. ...
Embodiment 1
[0260] Preparation of primers and photocleavable primers
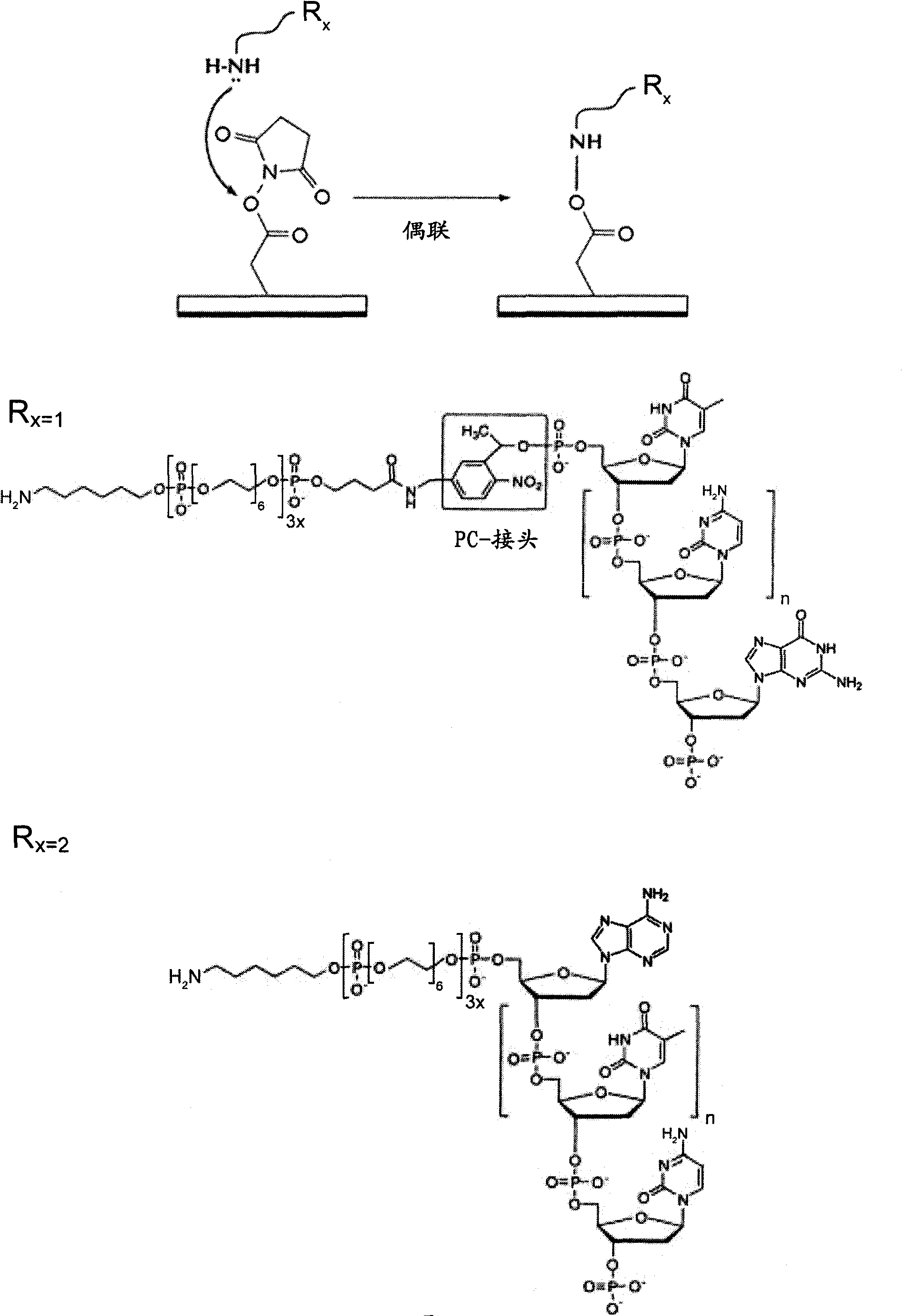
[0261] Oligonucleotide synthesis was performed on an ABI 394 synthesizer on a 4 x 1 μmol scale. Commercially available tacCPG (Proligo) was used as support material. All other chemicals used in standard synthetic reactions were obtained from Glen Research. Proligo's Phosphoramidite with a tert-butylphenoxy-acetyl protecting group (known as "tac" or "Expedite" monomer) was used. As the capping agent, tert-butylphenoxyacetylacetic anhydride (tac2O) in tetrahydrofuran was used.
[0262] Use the following commercially available modifiers:
[0263] -5' amino modifier C6: (6-(4-one methoxytritylamino)hexyl-(2-cyanoethyl)-(N,N-diisopropyl)-phosphoramidite
[0264] -Spacer phosphoramidite 18 (18-O-dimethoxytritylhexaethylene glycol, 1-[(2-cyanoethyl)-(N,N-diisopropyl)] - Phosphoramidite
[0265] - Photocleavable spacer [4-(4,4'-dimethoxytrityloxy)butyrylaminomethyl)-1-(2-nitrophenyl)-ethyl]-2- Cyanoethyl-(N,N-diisopropy...
Embodiment 2
[0281] Bead preparation and photolysis
[0282] Amino-modified oligonucleotides (Sequence ID #1-9) containing stationary and photocleavable linkers, respectively, were bound to N-hydroxysuccinamide ester (NHS) functionalized agarose beads according to standard methods (Roche / 454-Life Sciences, Branford, CT, USA). chemical reaction mechanism by figure 2 express.
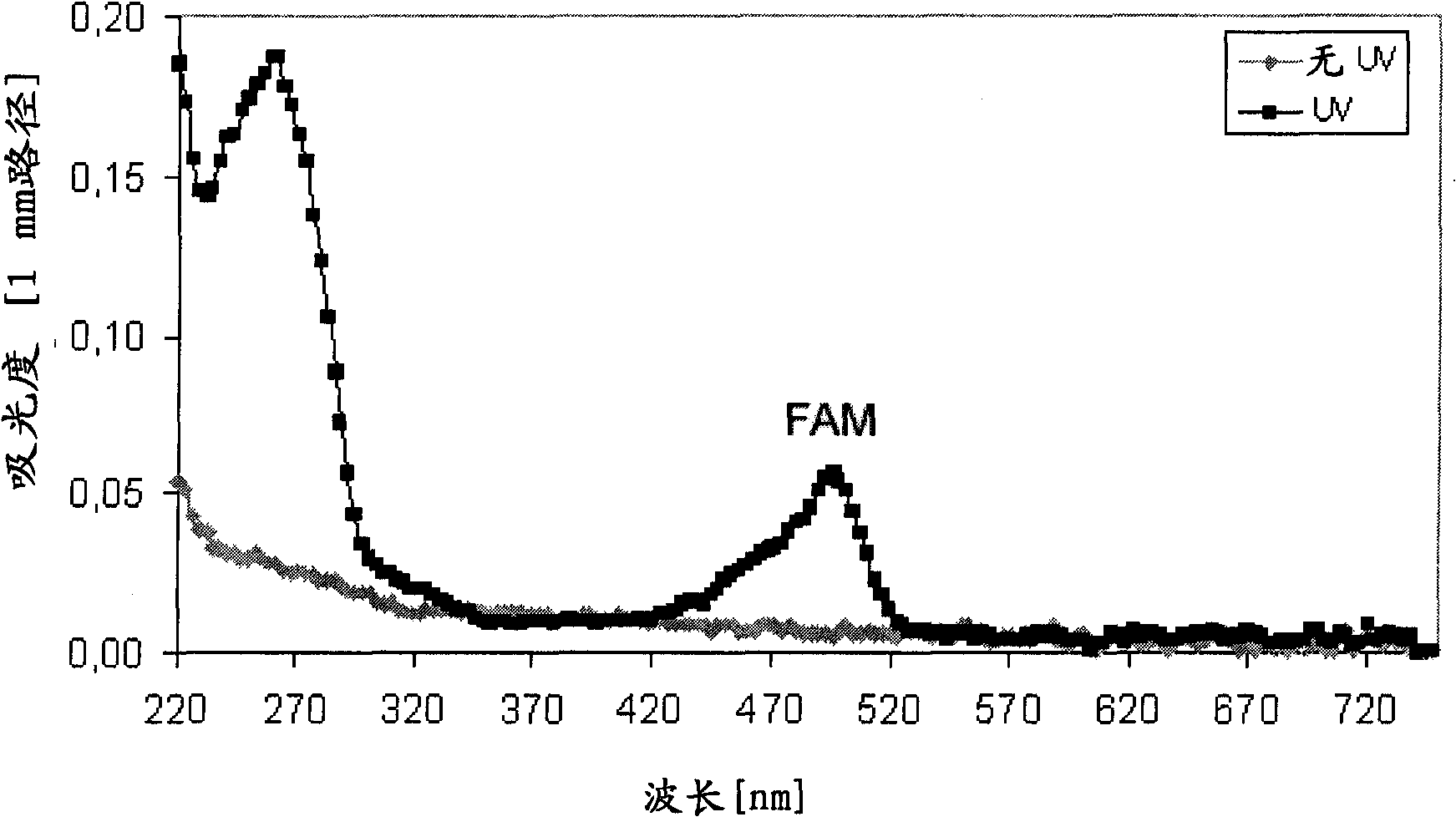
[0283] To trigger photocleavage of the nitrobenzyl linker, the beads were subjected to UV irradiation in a QS1.000 quartz cuvette (1-cm path length) using an 8W dual-wavelength UV lamp (Camag, Berlin, Germany). ) at 366nm. The distance between the quartz cuvette and the UV lamp is 2 cm.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com