Transgenosis construct and application thereof in preparing space-time adjustable liver damage model
A technology for liver damage and constructs, which is applied to cells modified by the introduction of foreign genetic material, introduction of foreign genetic material using vectors, animal husbandry, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0070] Animal model preparation
[0071] After obtaining the transgenic construct, conventional methods can be used to transfer the linearized construct into a fertilized egg. After the birth of the offspring animal, various methods known in the art (including but not limited to PCR detection, Southern blotting, etc.) can be used to identify whether the transgenic construct is integrated into its genome, thereby obtaining a transgenic animal.
[0072] Therefore, the present invention also provides a method for preparing a spatiotemporal adjustable liver injury model, the method comprising: (1) introducing the construct 1 of claim 1 into a fertilized egg of a non-human mammal, and transferring it into The fertilized egg with the construct is transferred into the fallopian tube of the pseudo-pregnant non-human mammal to continue its development to generate a non-human mammal integrated with the construct 1 of claim 1 in the genome; (2) claim 1 The construct 2 is introduced into a fe...
Embodiment 1
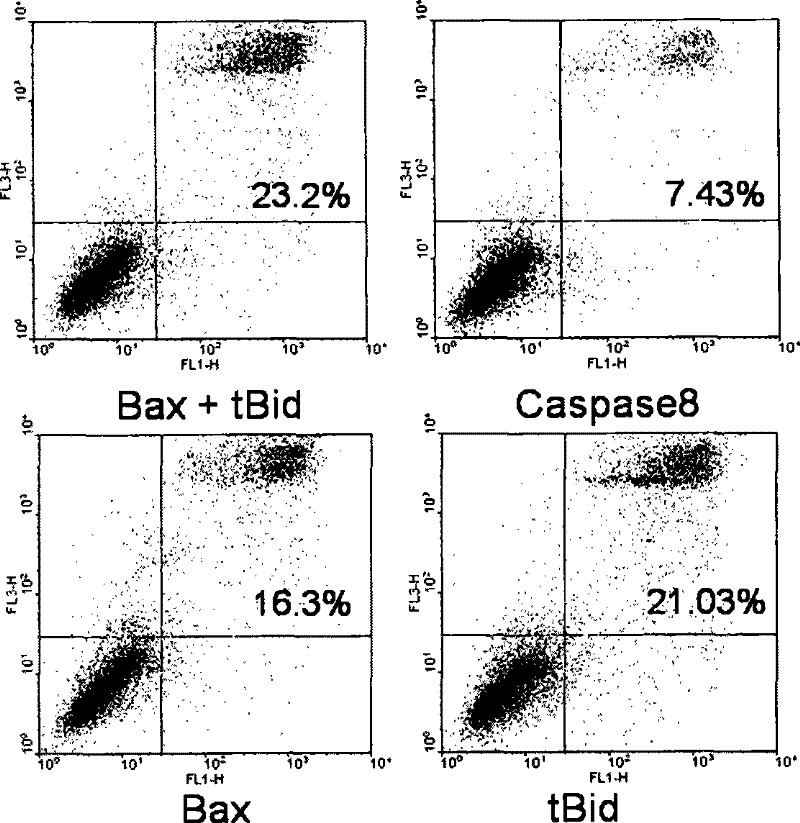
[0099] Example 1. Determine the most suitable gene for inducing hepatocyte apoptosis
[0100] It is known that there are many genes that can induce cell apoptosis. In this example, the most suitable gene for inducing hepatocyte apoptosis is first selected.
[0101] Using the conventional FACS method, the three genes Caspase8, Bax, tBid were compared to induce hepatocyte apoptosis to determine the most suitable gene. Caspase8, Bax, and tBid were cloned into the vector pcDNA3.1 (purchased from Invitrogen). Using mouse liver cDNA as a template, the primer sequences are (upstream primer sets Eco RI site, downstream primer sets XhoI site):
[0102] Caspase8F (SEQ ID NO: 2):
[0103] CTCT GAATTC CCATGGATTTCCAGAGTTGTCTTTATGC;
[0104] Caspase8R (SEQ ID NO: 3):
[0105] GCAC CTCGAG TTAGGGAGGGAAGAAGAGCTTCTT;
[0106] Bax F (SEQ ID NO: 4):
[0107] GCA GGAATTC CCATGGACGGGTCCGGGGAG;
[0108] Bax R (SEQ ID NO: 5):
[0109] GTGG CTCGAG TCAGCCCATCTTCTTCCAGATGG;
[0110] tBid F (SEQ ID NO: 6):
[0111]...
Embodiment 2
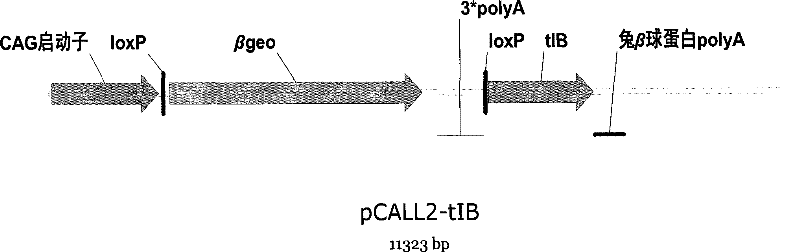
[0116] Example 2. Construction of Transgenic Vector
[0117] PCR was used to amplify the tBid and Bax genes from mouse liver cDNA and ligate them into the pMD18T-simple vector.
[0118] The primer sequence is as follows:
[0119] tBid F (SEQ ID NO: 8):
[0120] 5’-ATATAGATCTACCATGGGCAGCCAGGCCA-3’;
[0121] tBid R (SEQ ID NO: 7):
[0122] 5’-GCGCCTCGAGTCAGTCCATCTCGTTTCTAACCAAG-3’;
[0123] Bax F (SEQ ID NO: 9):
[0124] 5’-ATATAGATCTACCATGGACGGGTCCGGG-3’;
[0125] Bax R (SEQ ID NO: 5):
[0126] 5'-GTGGCTCGAGTCAGCCCATCTTCTTCCAGATGG-3'.
[0127] The PCR amplification conditions are as follows:
[0128] tBid: first 94°C, 5min; then 94°C, 30s; 56°C, 30s; 72°C, 30s; 30 cycles; finally 72°C, 10min.
[0129] Bax: first 94°C, 5min; then 94°C, 30s; 58°C, 30s; 72°C, 30s; 30 cycles; finally 72°C, 10min.
[0130] The IRES sequence was cut from pIRES2-EGFP with BglII and NcoI and ligated to the corresponding site in pET28a to obtain pET28a-IRES; then by PCR, Bax and tBid were amplified from liver cDNA and ins...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com