Phenol degrading and regulating gene and acting promoter thereof
A phenol degradation and promoter technology, applied in the field of promoters, can solve problems such as gene cloning and regulation mechanism that are not involved in the regulation of phenol degradation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
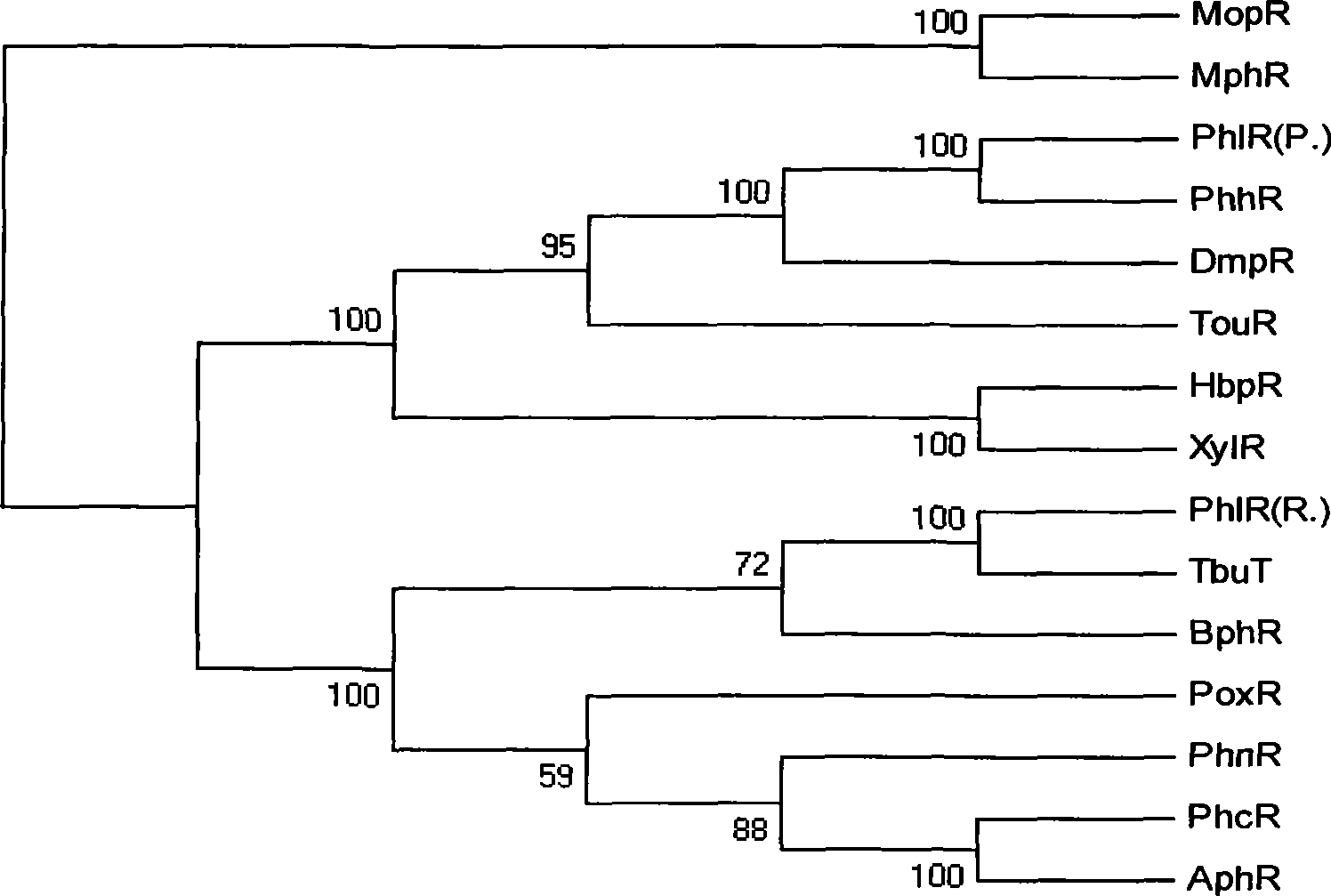
[0052] Example 1 Cloning and sequence analysis of the nucleotide sequence of SEQ ID NO: 1 in Acinetobacter calcoaceticus PHEA-2.
[0053] The present invention clones the sequence of SEQ ID NO: 1 located upstream of the phenol hydroxylase gene by reverse PCR method, and the specific process is as follows. Use EcoRI (Takala Company) to digest 5ug of Acinetobacter calcoaceticus PHEA-2 chromosomal DNA, digest the product T4 DNA ligase (NEB Company) at 10°C for overnight self-ligation, and design the reverse direction according to the cloned phenol hydroxylase gene sequence Primers, using the self-ligated product as a template to amplify a 3kb fragment, which was recovered and ligated into a T vector (Takala Company). Sequencing found that the cloned sequence extended the known phenol hydroxylase gene sequence upstream by 1961bp, including a complete open reading frame of 1671bp, encoding 556 amino acids. The protein SEQ ID NO: 3 encoded by sequence SEQ ID NO: 1 is found to be a ...
Embodiment 2
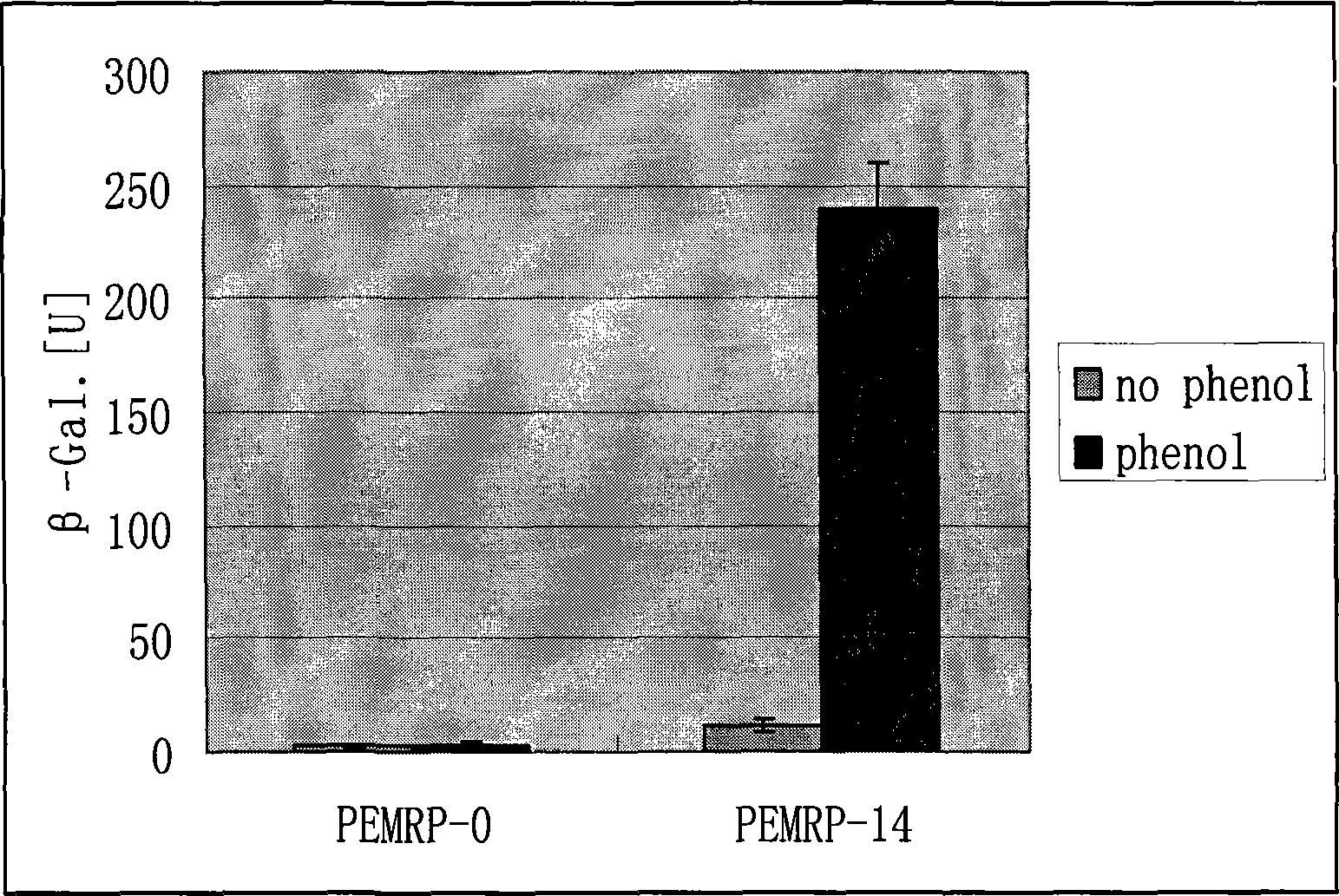
[0054] Example 2 Expression of SEQ ID NO: 3 polypeptide in Escherichia coli and construction of whole cell biosensor.
[0055] (1) Construction of the prokaryotic expression vector of SEQ ID NO:1.
[0056] The two ends of the sequence of SEQ ID NO: 1 were introduced into the NdeI and SalI restriction sites by PCR method, and the amplified sequence was digested with NdeI and SalI (Takala Company) and then ligated with the same expression vector pET28a (NEB Company) The recombinant expression vector pEMR was obtained.
[0057] (2) Construct the test expression vector of SEQ ID NO:2.
[0058] The two ends of the sequence of SEQ ID NO: 2 were introduced into XhoI and PstI restriction sites by PCR method. The amplified sequence was digested with XhoI and PstI (Takala Company) and then connected to the upstream of the lacZ gene without a promoter of the promoter detection vector pPR9Tt (Pedro Miguel Santos, 2001) digested with the same enzymes, and the phenol hydroxylase promoter ...
Embodiment 3
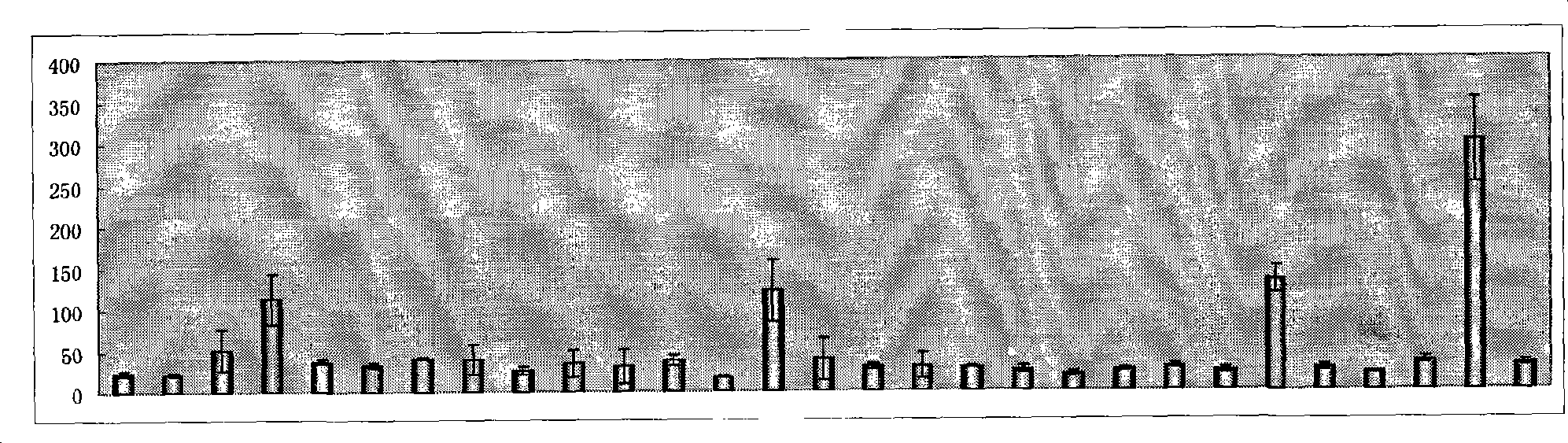
[0061] Example 3 Co-transformation bacteria PEMRP-14 sensed the range of phenolic compounds.
[0062]Using 28 different phenolic compounds as inducers to induce PEMRP-14 bacteria, and measuring the activity of β-galactosidase, it was found that phenol, catechol, o-cresol and 2-chlorophenol can be encoded by SEQ ID NO: 1 Protein recognition ( image 3 ).
[0063] The specific method is as follows: pick freshly cultivated colonies, shake the bacteria at 37 degrees overnight, and insert 1% inoculum into 29 test tubes of LB medium the next day, shake until the OD600 reaches about 0.6, and insert different phenols into each tube The compound, with a final concentration of 0.3mM, was incubated for 2 hours, and 29 different samples were taken out to measure the enzyme activity of β-galactosidase. The determination method was referred to "Molecular Cloning" (Sambrook et al., 1989). The results showed that MphR could recognize four compounds, phenol, catechol, o-cresol and 2-chloroph...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com