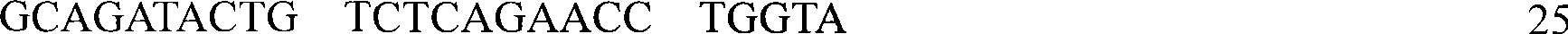CXCL10 gene polymorphism relevant to progression of chronic hepatitis type B disease course and detection method thereof
A chronic hepatitis B, gene technology, applied in the genotype detection of the susceptibility gene CXCL10, the detection kits for the genotypes of the susceptibility genotypes mentioned above, can solve the problem of changing expression or binding activity, affecting liver damage and disease severity, etc. problems to reduce the risk of occurrence
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] In this example, a set of primer pairs was designed and synthesized, and based on the primer pairs, a method for detecting the G-201A genotype of the CXCL10 gene associated with the progression of hepatitis B was designed.
[0027] 1. Genomic DNA extraction
[0028] Genomic DNA of peripheral blood leukocytes was extracted by the phenol / chloroform method in the field.
[0029] 1) Take sodium citrate or EDTA-Na 2 5ml of anticoagulated whole blood (try not to use heparin for anticoagulation), put in a 50ml capped centrifuge tube;
[0030] 2) Add 3-5 times the volume of cold distilled water, invert and mix repeatedly, place in ice for 5 minutes, and centrifuge at 2000rpm at 4°C for 20 minutes;
[0031] 3) Slowly decant the supernatant, add pre-cooled 0.1% Triton-X100 (same volume as above) to the precipitate, gently mix the precipitate, centrifuge as above, and discard the supernatant;
[0032]4) Add 4ml of lysate (50Mm Tris-Cl-10mM EDTA, pH 8.0) to the precipitate, brea...
Embodiment 2
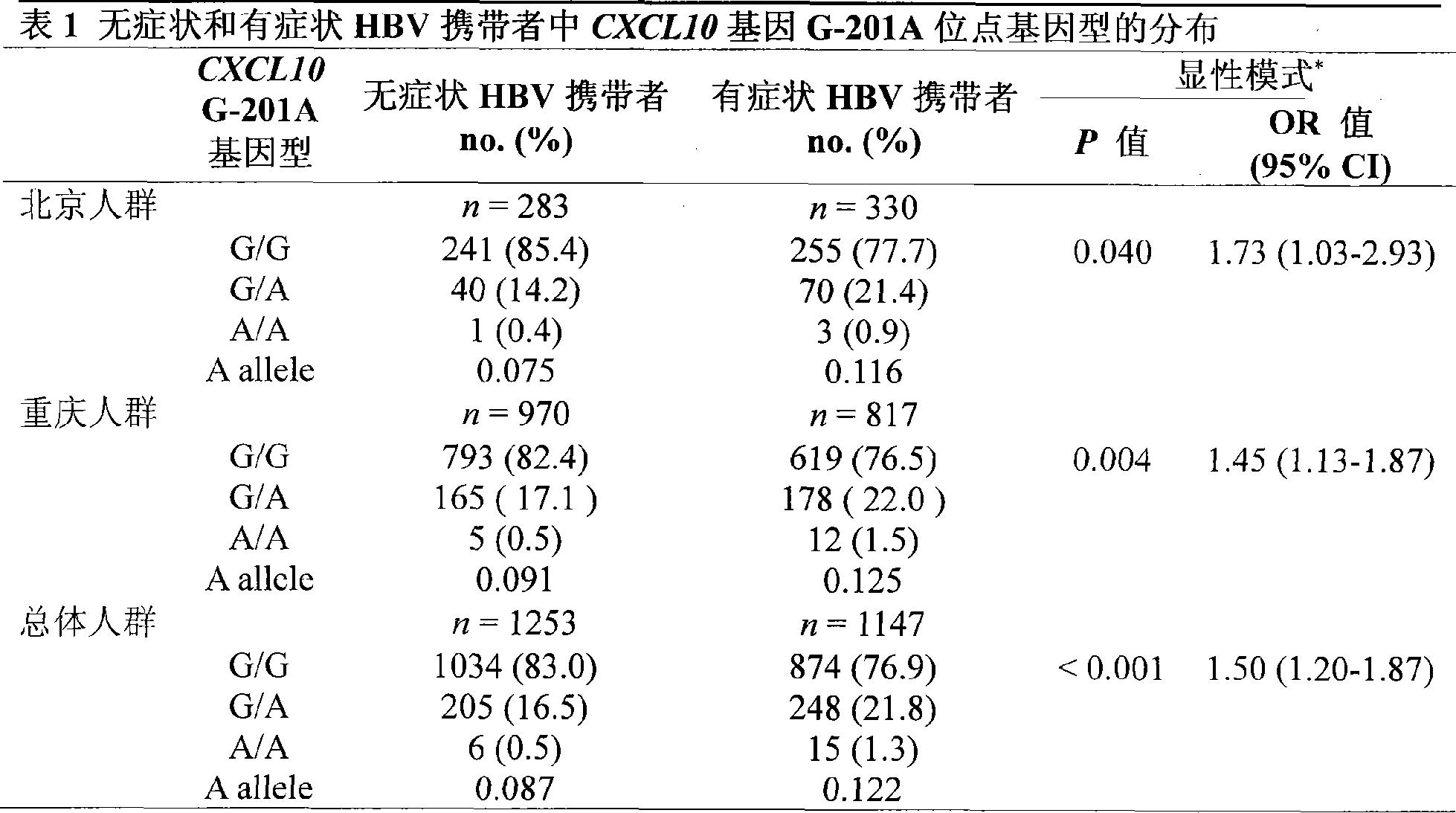
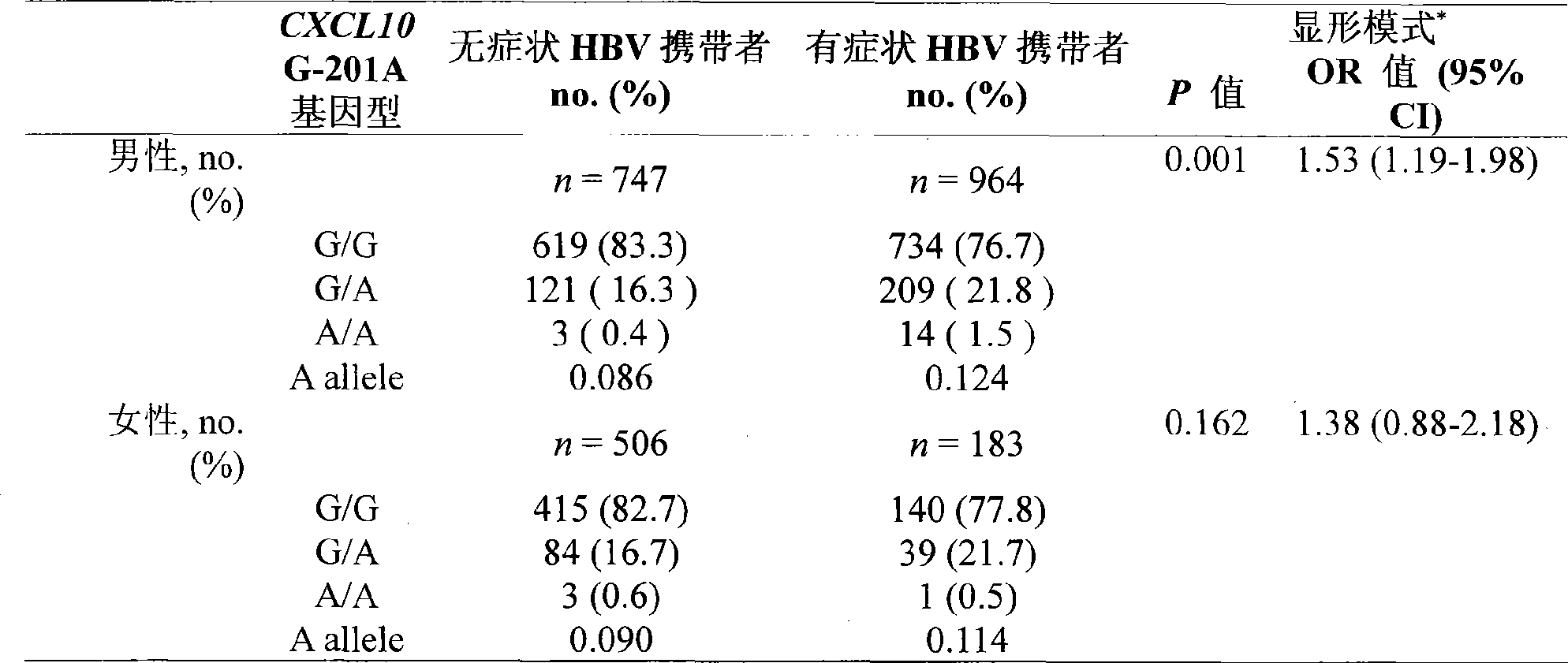
[0046] In this example, the method established in Example 1 was used to analyze the G-201A genotype of the CXCL10 gene of 1147 cases of chronic progressive hepatitis B patients and 1253 cases of asymptomatic HBV carriers.
[0047] 1. Test object:
[0048] This study included 1147 chronic progressive hepatitis B patients and 1253 asymptomatic HBV carriers (collected from Beijing Hospital of Infectious Diseases [613 copies] and Chongqing Southwest Hospital [1787 copies] from February 2001 to March 2006). ]). All HBV-infected patients were positive for HBsAg and anti-HBc IgG for at least 12 months. All carriers underwent liver function test, serum marker detection, ultrasound / computed tomography imaging during the study period, and 376 cases (15.7%) underwent liver biopsy. All subjects had no serological evidence of HCV, HDV, and HIV co-infection. While collecting tissue specimens or blood samples, each research subject signed an informed consent form and provided detailed cli...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com