Multiple PCR detecting reagent case for diagnosis of oesophagostomiasis in pig
A detection kit and technology for nematode disease, applied in the direction of microbial determination/inspection, biochemical equipment and methods, etc., to achieve simple operation, high sensitivity, and objective result judgment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0056] Example 1 Composition of the kit
[0057] The kit contains 30ml DNA lysis solution, which contains 100mM NaCl, pH8.0 10mM Tris-Cl, pH8.0 25mM EDTA, 1% (W / V) SDS and 1.7μg / μL proteinase K ; PCR reaction solution 100 reactions (25μL / reaction), final concentration of each 200μM dATP, dTTP, dGTP, dCTP, final concentration of each 0.5pmol / μL primers OdspF, OdspR2, OqspF and OqspR, final concentration of 3.5mM MgCl 2 , Taq enzyme 25μL (5U / μL); one for each positive control of dentate esophageal nematode DNA and one positive control of four-spinned esophageal nematode.
Embodiment 2
[0058] Example 2 Kit specificity test
[0059] Using 5 control samples of Ascaris suum, Trichinella spiralis, Trichinella, Clonorchis sinensis, and Schistosoma japonicum that have been validated by DNA validity, 1μL each of the DNA was used as a template to perform specific PCR amplification according to the reaction conditions of the kit. Blank control and kit positive control.
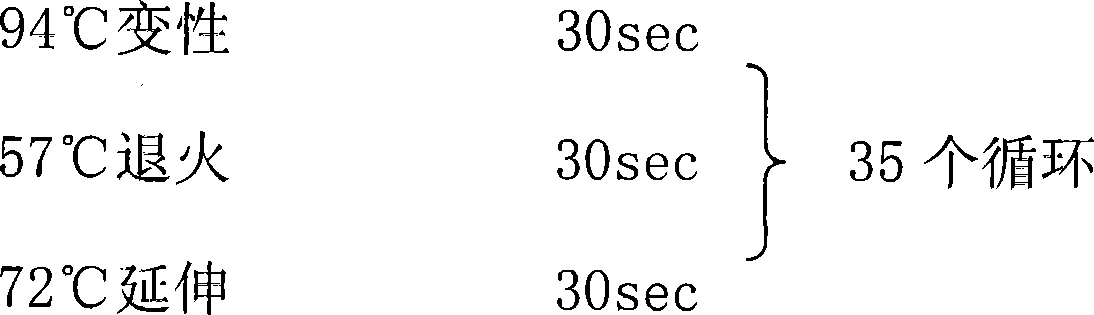
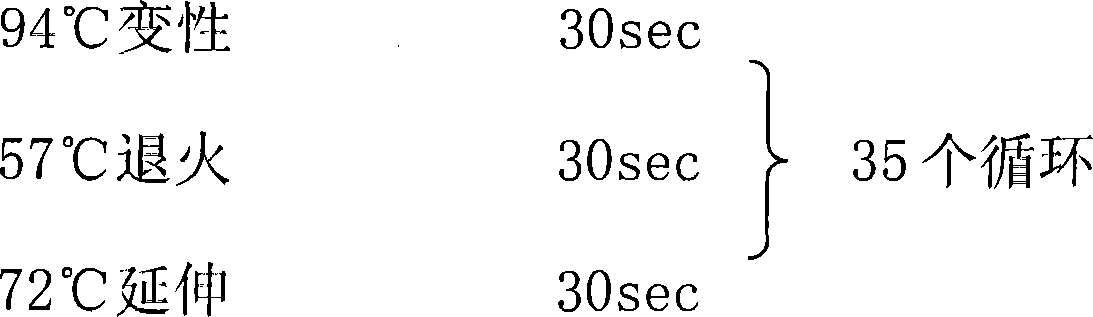
[0060] PCR amplification conditions: 94℃ pre-denaturation 5min
[0061]
[0062] Extension 5min after 72℃
[0063] After the PCR products were electrophoresed in a 1.2% TBE agarose gel, the results were observed under a UV transilluminator and photographed by a gel imaging system.
[0064] Results The kit has a DNA positive control of esophageal esophagus nematode and amplifies a band of about 130 bp. The positive control of four spines esophageal nematode DNA amplifies a band of about 330 bp. The mixed DNA of the two positive controls amplifies a band of about 130 bp and The band of 330b...
Embodiment 3
[0065] Example 3 Sensitivity test of the kit
[0066] First, the dentate esophageal mouth nematode sample and the four-spinned esophageal mouth sample nematode extract DNA, then dilute, vortex and mix, and detect the total DNA content according to the operating procedures of the Eppendorf Biophotometer nucleic acid protein analyzer. Dilute DNA by 5×, 10×, 20×, 50×, 100×, 200×, 400×. The PCR amplification conditions are the same as above, and a blank control is set. The PCR product was detected by 1.2% agarose gel electrophoresis to determine its sensitivity. The test results show that the PCR detection method is highly sensitive, and can detect 0.1ng DNA dentate esophagus nematodes and 0.12ng DNA four spines esophagus nematodes at the lowest.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com