Construction method for producing 1,3-trimethylene glycol regrouping saccharomyces cerevisiae with glucose as substrate
A technology for recombining Saccharomyces cerevisiae and propylene glycol, applied in the field of genetic engineering, can solve the problems of increasing fermentation cost and high price
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
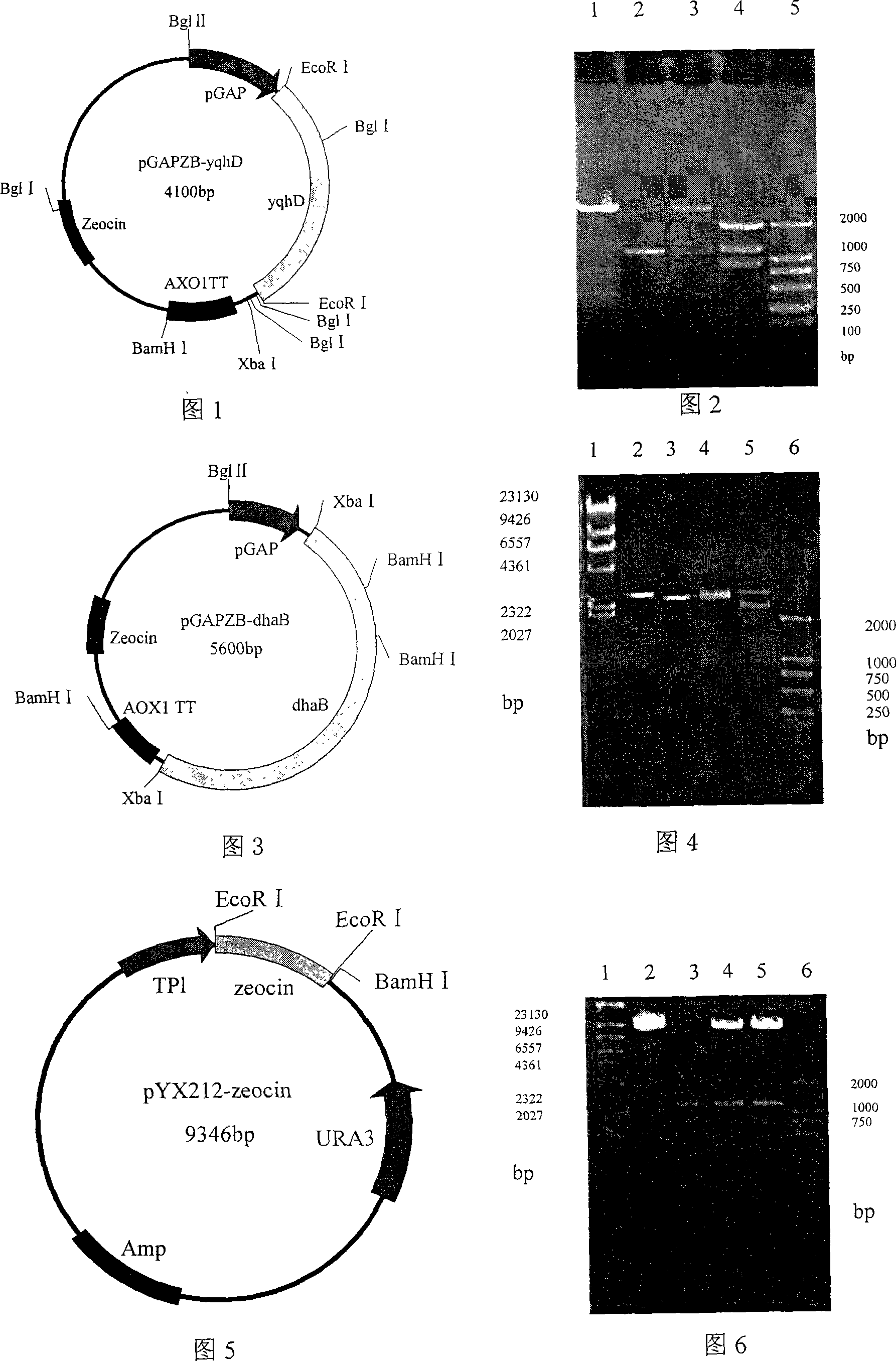
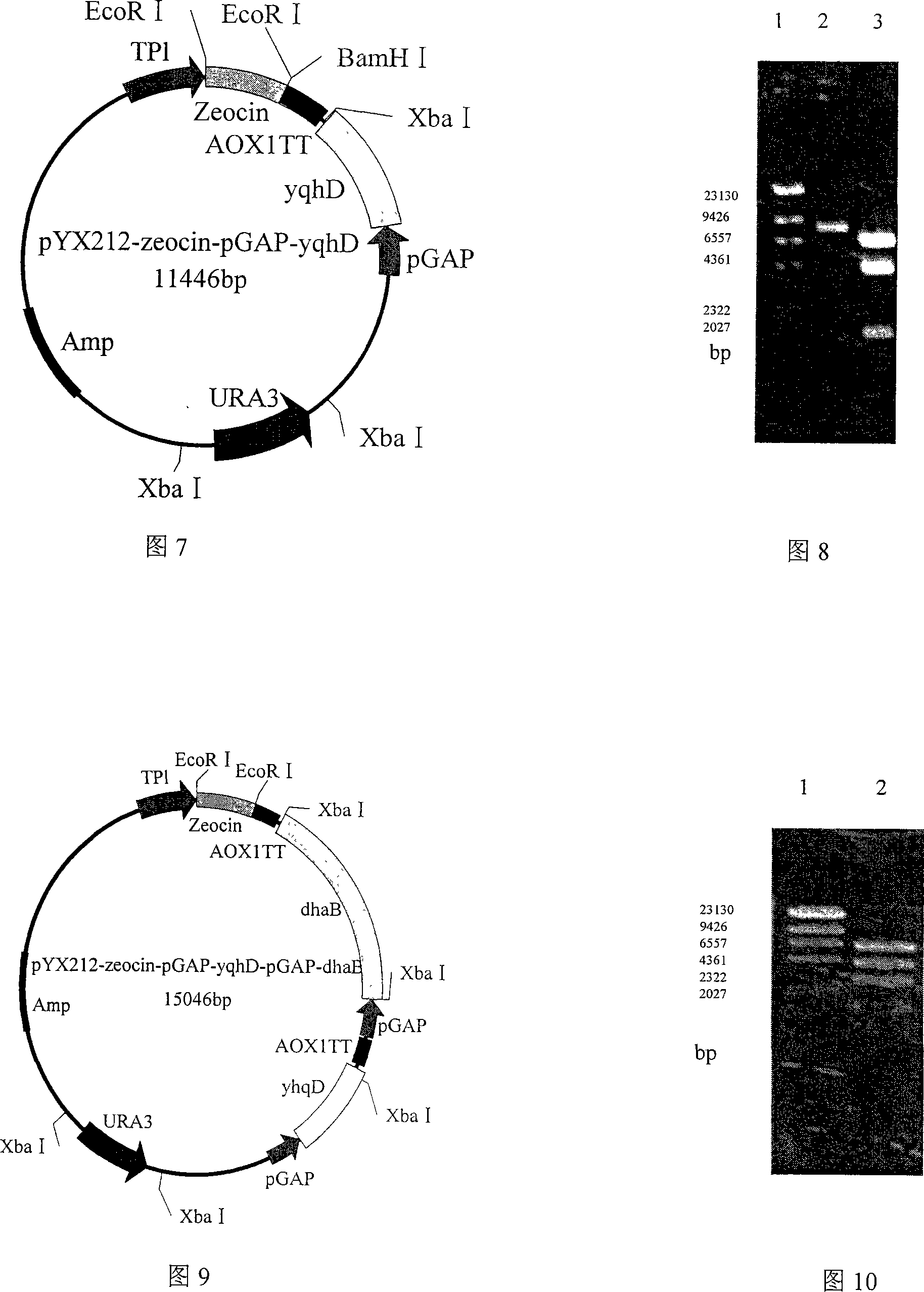
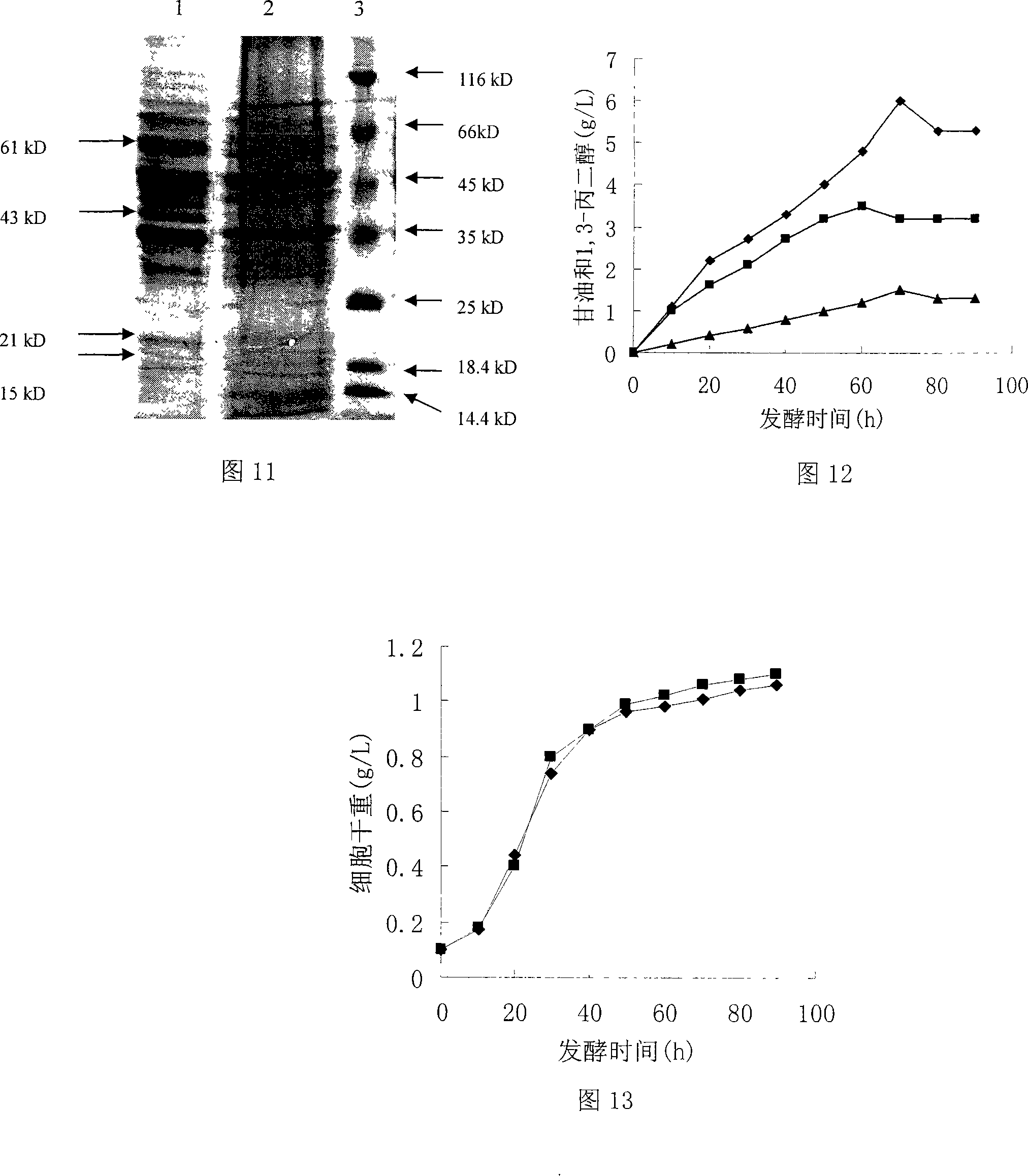
[0057] Embodiment 1: Construction of plasmid PGAPZB-yqhD:
[0058] EcoR I cuts the vector pHsh-dhaB-yqhD, and releases the 1,3-propanediol oxidoreductase isoenzyme coding gene yqhD with a size of about 1.2kb, which is purified with a PCR fragment gel recovery kit; extracts the plasmid PGAPZB, and uses EcoR I digest the vector PGAPZB, purify by agarose gel electrophoresis, reclaim the 2.9kb target fragment with the PCR fragment recovery kit, and use T 4 Ligase ligated yqhD / EcoR I and PGAPZB / EcoR I, and the ligation product was transformed into Escherichia coli JM109 competent cells, and the transformed product was spread on LB agar plate containing 25 μg / mL Zeocin resistance, and cultured overnight at 37°C; 5 colonies were randomly selected, Inoculate in 20 mL of LB medium containing 25 μg / mL Zeocin resistance and culture overnight at 37°C, extract plasmids, digest with EcoR I and Bgl I respectively, identify positive clones by agarose gel electrophoresis, and store positive re...
Embodiment 2
[0059] Embodiment 2: Construction of plasmid PGAPZB-dhaB:
[0060] The vector pHsh-dhaB-yqhD was digested with Xba I to release the glycerol dehydratase coding gene dhaB with a size of about 2.7kb, which was purified with a PCR fragment gel recovery kit; the plasmid PGAPZB was extracted, the vector PGAPZB was digested with Xba I, and gelatinized in agarose Purified by gel electrophoresis, recovered a 2.9kb target fragment with a PCR fragment recovery kit, and used T 4 Ligase ligated dhaB / Xba I and PGAPZB / Xba I, and the ligation product was transformed into Escherichia coli JM109 competent cells, and the transformed product was spread on LB agar plates containing 25 μg / mL Zeocin resistance, and cultured overnight at 37°C; 5 colonies were randomly selected , inoculated in 20mL LB medium containing 25μg / mL Zeocin resistance and cultured overnight at 37°C, extracted plasmids, digested with Xba I and BamH I respectively, and identified positive clones by agarose gel electrophoresis...
Embodiment 3
[0061] Embodiment 3: Construction of plasmid pYX212-zeocin:
[0062] Using the vector PGAPZB as a template, the resistance gene zeocin with a size of 1.2kb was amplified by PCR, and purified with a PCR fragment gel recovery kit; the plasmid pYX212 was extracted, and the vector pYX212 was digested with EcoR I, purified by agarose gel electrophoresis, and purified with PCR fragments The recovery kit recovers the target fragment of 8.3kb, using T 4 Ligase zeocin / EcoR I and pYX212 / EcoR I, and the ligation product was transformed into Escherichia coli JM109 competent cells, and the transformed product was spread on LB agar plate containing 25 μg / mL Zeocin resistance, cultured overnight at 37°C; 5 colonies were randomly selected , inoculated in 20mL LB medium containing 25μg / mL Zeocin resistance and cultured overnight at 37°C, extracted plasmids, digested with EcoR I respectively, identified positive clones by agarose gel electrophoresis, and stored positive recombinants in a medium...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com