Porphyra haitanensis germ plasma identification fingerprint construction method and its uses
A technology of fingerprint spectrum and construction method, applied in the field of fingerprint spectrum construction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
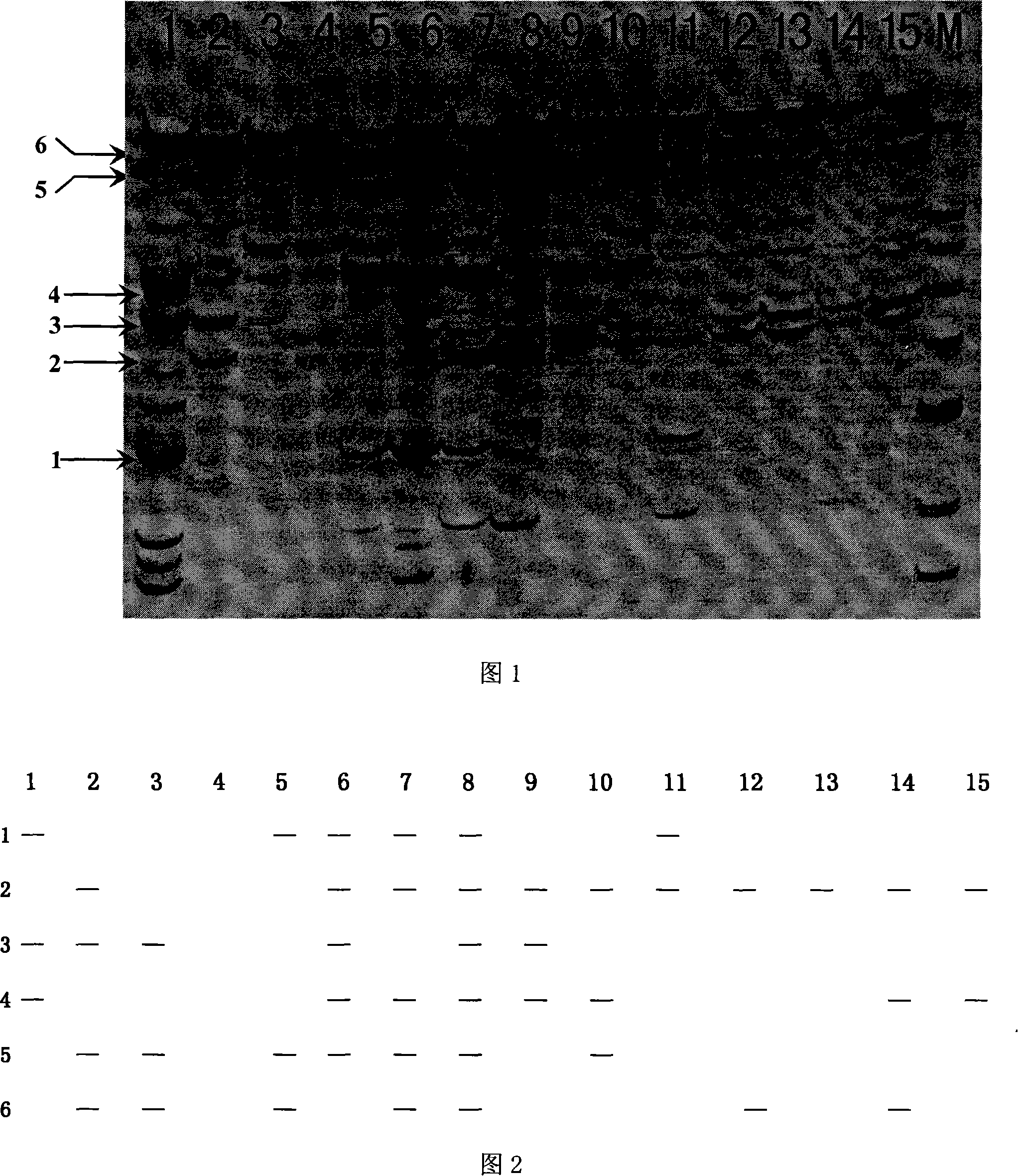
[0033] In this example, the collected thallus of 15 Porphyra laver strains that are widely representative in production or research are used as test materials, the total DNA of the genome is extracted, the SRAP molecular marker analysis of the specified primers is carried out, and the representative ones are selected. Polymorphic bands, establish their DNA fingerprints, and convert the fingerprints into digital fingerprints composed of "0" and "l", input the fingerprint recognition software to establish the DNA digital fingerprint library of almond laver, and finally construct the The DNA digital fingerprint library was used in the germplasm identification of Porphyra alba. The specific implementation method is as follows:
[0034] 1. Selected experimental materials:
[0035] Select the Porphyra laver line that is widely used in partial production or research, as shown in Table 1, the Porphyra laver strain that is widely used in production or research that has been collected ...
Embodiment 2
[0072] Apply the altar laver digital fingerprint library constructed in the present invention to the germplasm identification of altar laver:
[0073] Assuming that it is necessary to determine which algae strain a batch of almond laver belongs to which has been used on a large scale in production, this batch of altar laver can be identified according to the following method, and finally determine which strain it belongs to.
[0074] First, the genomic DNA was extracted from this batch of laver according to the method in Example 1, and purified as a template, using ME1: 5'-TAGGTCCAAACCGGATA-3' as the forward primer, and EM9: 5'-GACTGCGTACGAATTAAC-3' as the reverse primer , using the SRAP amplification experimental system for SRAP amplification, the amplified product was electrophoresed on 8% polyacrylamide gel at 60W constant power for 1.5h, and was visualized by silver staining, according to the selected 6 polymorphic bands Draw the DNA fingerprints of each sample of this bat...
Embodiment 3
[0077] In this embodiment, 21 Porphyra laver strains (YSI~YSIV, YSV-①~YSV-③, YSVI, YSVII-①~YSVII-⑤, YSX-①~YSX-⑤ and YSXI~YSXIII) thallus as the test material, extract the total DNA of the genome, carry out SRAP molecular marker analysis with specified primers, select representative polymorphic bands, establish their DNA fingerprints, and convert the fingerprints into For the digital fingerprint composed of "0" and "1", input the fingerprint recognition software to build the DNA digital fingerprint library of Porphyra alba, and finally use the constructed DNA digital fingerprint library in the germplasm identification of Porphyra almanac. The specific implementation method is as follows:
[0078] 2. Selected experimental materials:
[0079] Twenty-one Porphyra laver strains (YSI~YSIV, YSV-①~YSV-③, YSVI, YSVII-①~YSVII-⑤, YSX-①~YSX-⑤ and YSXI~YSXIII) in the Fujian Porphyra Germplasm Bank were used. ) fronds.
[0080] 2. Extraction and purification of Porphyra japonica genomic ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com