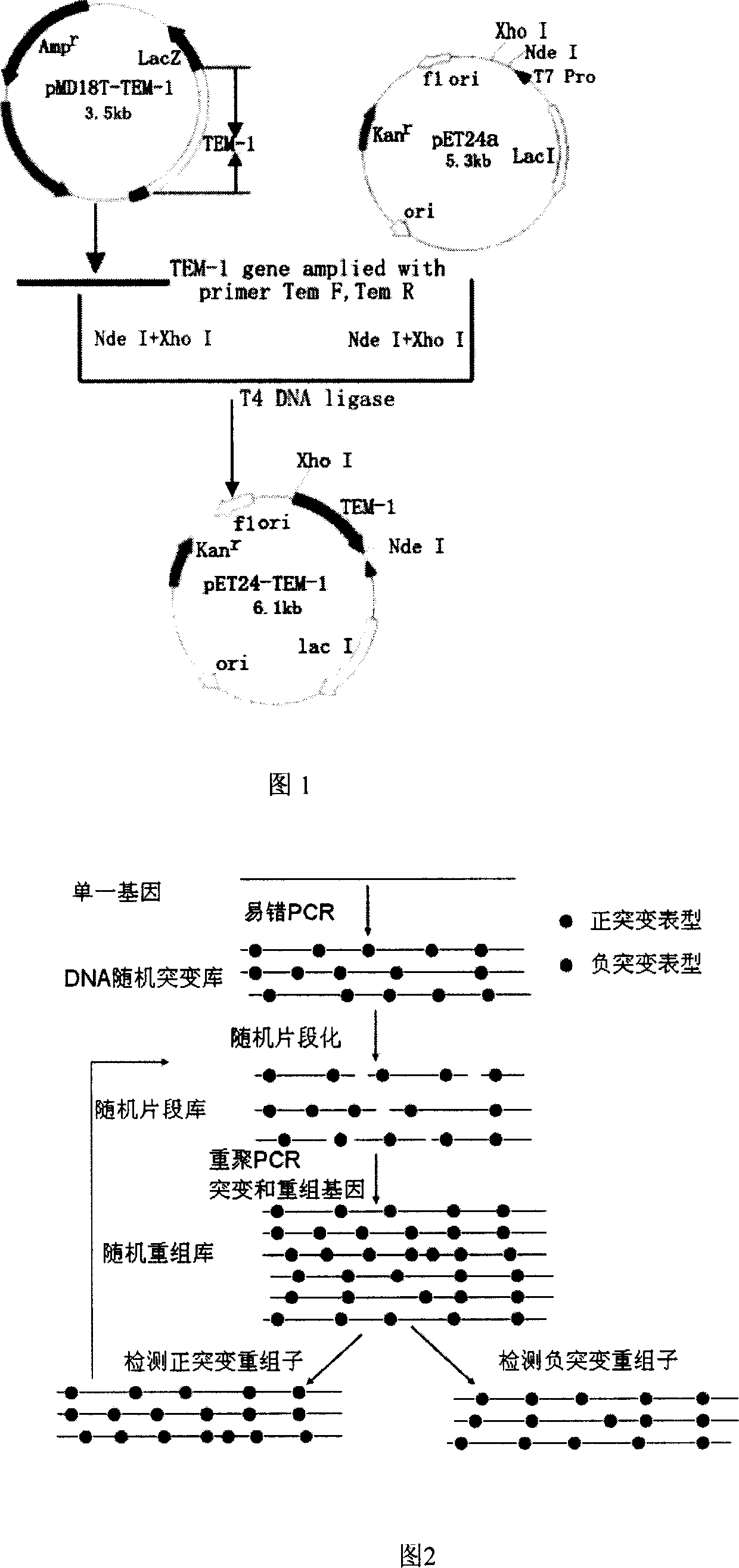
Method for obtaining high-vitality superspectrum beta-lactam enzyme by directional anagenesis in vitro
A technology of lactamase and high activity, applied in the field of medical bioengineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Embodiment 1: Cloning of TEM-1 gene
[0030] (1) PCR amplification of TEM-1 fragment
[0031] Using the TEM-1 gene as a template, design primers and add enzyme cutting sites. The above-mentioned enzyme-producing E.coli plasmid was extracted and used as a template for PCR reaction. The reaction conditions were: pre-denaturation at 94°C for 4 min, followed by 20 cycles with the following parameters: denaturation at 94°C for 1 min, annealing at 58°C for 50 s, extension at 72°C for 1 min, and the last cycle of extension at 72°C for 10 min.
[0032] (2) Purification of PCR products
[0033] The low melting point agarose (Low melting point agarose, LMPA) method was used for recovery. After the PCR product was subjected to 1% low-melting point agarose gel electrophoresis, the gel piece containing the target fragment was cut out, put into a 1.5ml Eppendorf tube, and about 200 μl of TE was added, and an equal volume of Tris saturated phenol was added to melt at 70°C for 10 C...
Embodiment 2
[0046] Embodiment 2: TEM-1 gene error-prone PCR
[0047] Error prone PCR:
[0048] 10× error-prone PCR buffer: containing 4.4mmoL / L MgCl2, 500mmoL / L KCl, 100mmoL / LTris-HCl (pH7.4), 0.1% (w / v) gelatin.
[0049] 10×dNTP mixture: contains 2mmoL / L dGTP, 2mmoL / L dATP, 10mmoL / L dCTP, 10mmoL / L dTTP.
[0050] 5mmoL / L MnCl2 solution.
[0051] Reaction system: volume
[0052] 10×Taq DNA polymerase 1μl
[0053] 10× error-prone PCR buffer 10μl
[0054] 10×dNTP mix 10μl
[0055] dCTP, dTTP 100mM 1μl each
[0056] Template DNA 4μl
[0057] Upstream primer 1μl
[0058] Downstream primer 1μl
[0059] Ultrapure water 51μl
[0060] 2.5mmol / L MgCl 2 10μl
[0061] 5mmoL / L MnCl 2 10μl
[0062] Total 100μl
[0063] PCR reaction conditions: time
[0064] Melting temperature 95°C 1min
[0065] Extension temperature 50℃ 1min
[0066] Annealing temperature 72℃ 3min
[0067] React 40 cycles,
[0068] Annealing temperature 72℃ 10min
[0...
Embodiment 3
[0070] Embodiment 3: Construction and screening of error-prone PCR mutation library
[0071] (1) Purification of mismatched PCR products is the same as the second step in Example 1
[0072] (2) Restriction enzyme digestion of mismatched PCR products
[0073] Digested with restriction enzymes Nde I and Xho I
[0074] Purify mismatched PCR product 8μl
[0075] XhoI 1μl
[0076] Nde I 1μl
[0077] 10×Universal Buffer 2μl
[0078] Deionized water 8μl
[0079] Total 20μl
[0080] Mix and incubate at 37° C. for 1 hour, and perform 2% agarose gel electrophoresis.
[0081] (3) Restriction digestion of pET24a vector DNA
[0082] Digest the plasmid vector with restriction enzymes Nde I and Xho I:
[0083] Plasmid DNA 4μl (about 2μg)
[0084] Nde I 1μl
[0085] XhoI 1μl
[0086] 10×Universal Buffer 2μl
[0087] Deionized water 12μl
[0088] Mix and incubate at 37°C for 1 hour, total 20μl
[0089] Perform 1% agarose gel electrophoresis to detect ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com