Novel eukaryotic cells and methods for recombinantly expressing a product of interest
a technology of eukaryotic cells and products, applied in the field of recombinant expression technologies, can solve the problems of loss of recombinant protein expression in cell clones, instability seriously affecting the industrial production process of recombinantly produced polypeptides, and expression yield declin
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
C12orf35 Gene Expression by RNA Interference (RNAi) in CHO Cells Increases Expression Yield
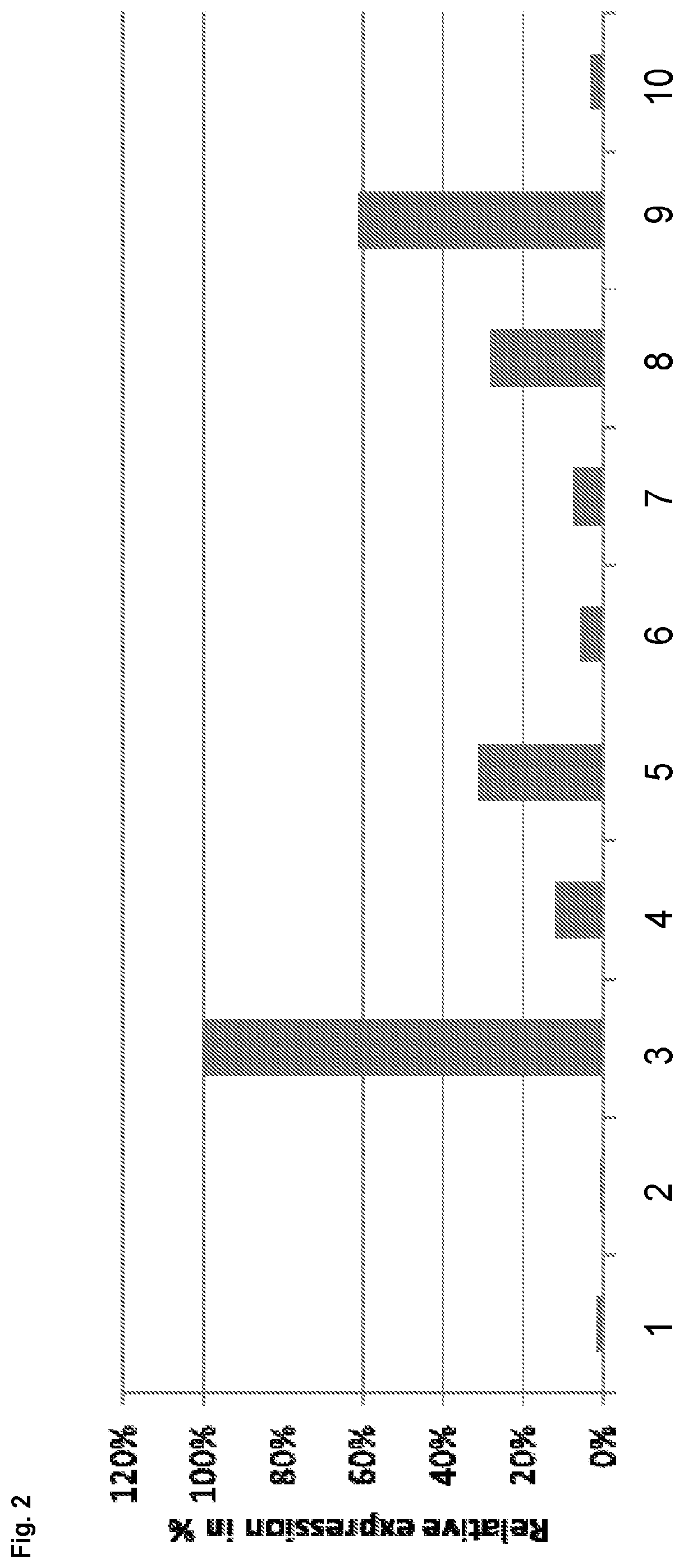
[0181]In order to demonstrate that reducing expression of gene C12orf35 results in an increase in volumetric or specific productivity, siRNAs were designed against different target genes located in the telomeric region of chromosome 8 of the Chinese hamster genome analysed (CHO-K1). siRNAs were designed against the following target genes listed in Table 2:
TABLE 2siRNAs against different target genesTarget geneSenseAntisenseMETTL20_1CCCUGAUGUUGUUAGAGGATTUCCUCUAACAACAUCAGGGTT(4833442J19Rik)(SEQ ID NO: 23)(SEQ ID NO: 24)C12orf35_1CAUCCAGACAAAUCUUACATTUGUAAGAUUUGUCUGGAUGTG(SEQ ID NO: 25)(SEQ ID NO: 26)C12orf35_2CCAGAAAGAUAAAUCUACATTUGUAGAUUUAUCUUUCUGGTA(SEQ ID NO: 27)(SEQ ID NO: 28)Caprin2_6UGACCUGCCCUGAAAGAAATTUUUCUUUCAGGGCAGGUCAGT(SEQ ID NO: 29)(SEQ ID NO: 30)FAM60AGCUUCCAGCUCUAACAGAATTUUCUGUUAGAGCUGGAAGCCA(SEQ ID NO: 31)(SEQ ID NO: 32)IPO8_1GACCCGAACUUUGACCCUATTUAGGGUCAAAGUUCGGGUCTG(SEQ ID NO: ...
example 2
n of a CHO Cell Line Which Comprises a Deletion in the Telomeric Region of Chromosome 8
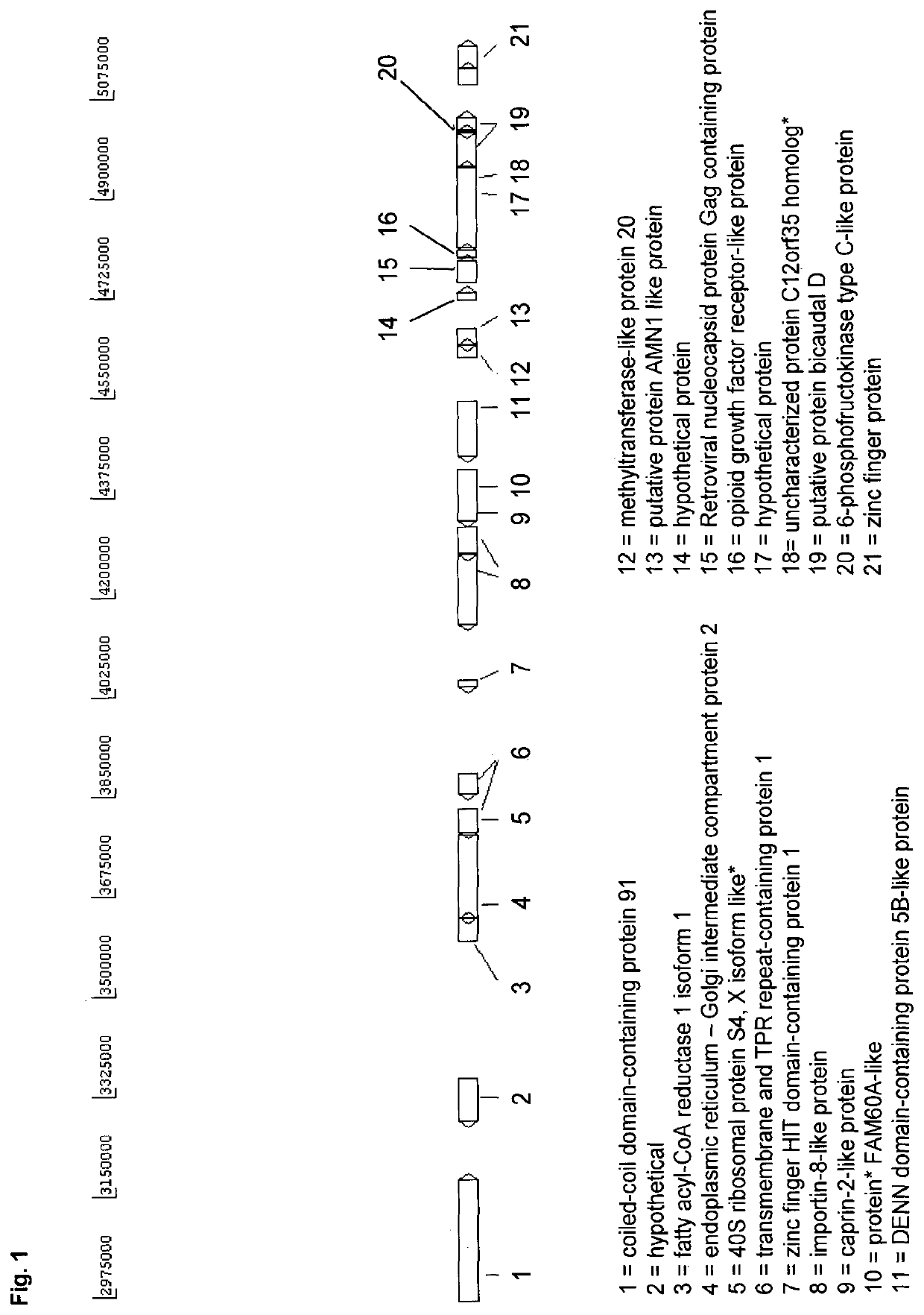
[0190]A novel CHO cell line (C8DEL) was generated, which comprises a deletion in the telomeric region of the q arm of chromosome 8. The deletion was induced by chromosome breakage. The deleted portion comprised gene C12orf35 as well as among others gene FAM60A (see FIG. 1). Said novel cell line was obtained from a parental cell line derived from CHO-K1. Said cell line was prepared as follows. The parental CHO cells were split at 2E5 cells / ml in culture medium comprising 0.5 μM, 1 μM or 2 μM MTX. After six days the cell viabilities were around 30-40%. Cells were centrifuged at 180×g for 5 min and cultivated in culture medium without MTX to allow the cells to recover until viabilities were above 95% (after ca. 21 days). This procedure was repeated two more times. Single cell clones were obtained from cell pools. Overall 561 cell clones were grown and DNA was isolated using the “Extract-N-Amp Blood P...
example 3
of the Characteristics of the Cell Line C8DEL
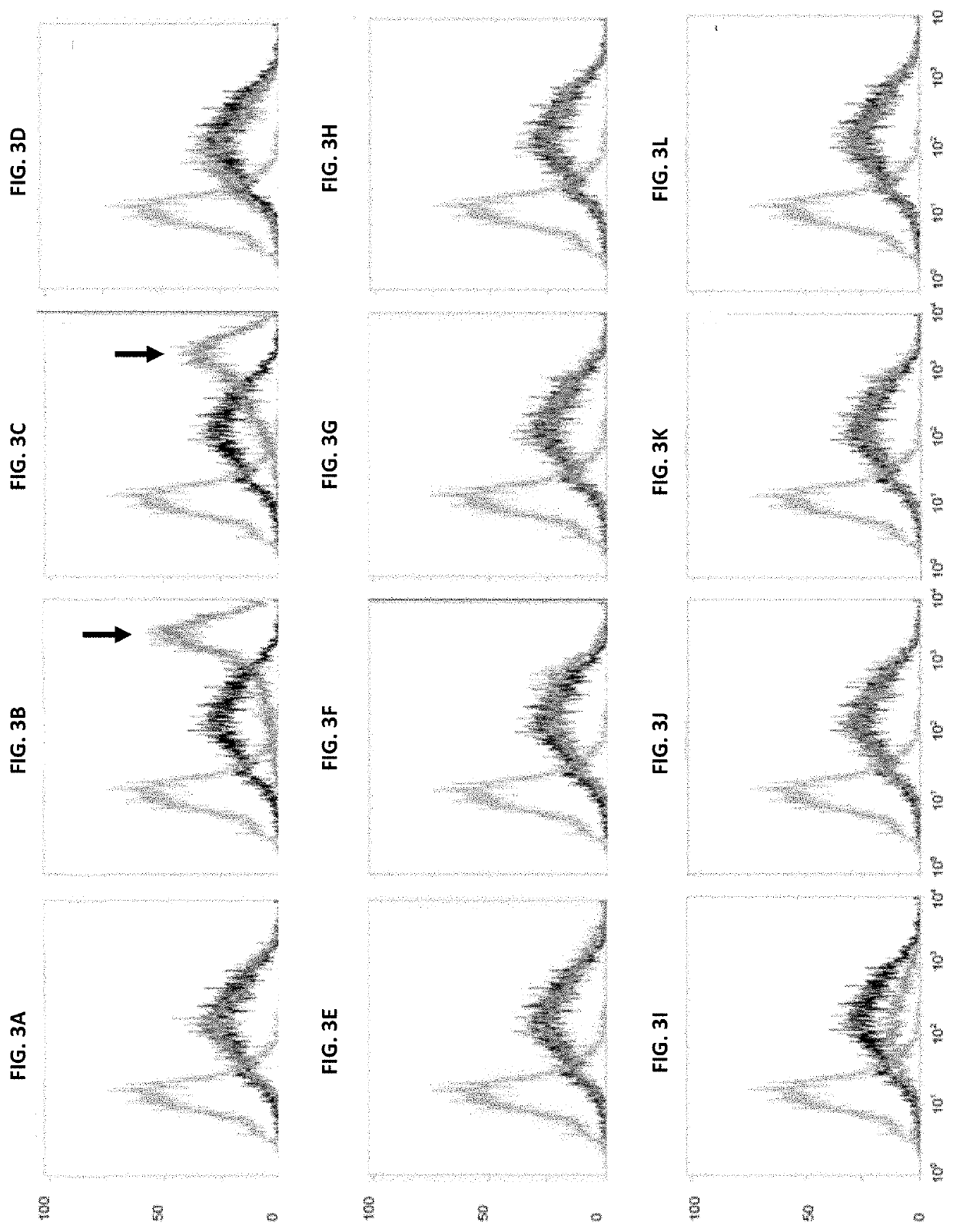
[0192]The cell line C8DEL was analyzed for its performance when recombinantly expressing a polypeptide of interest and compared to the parental cell line from which the cell line C8DEL was derived. As described above, said parental cell line does not comprise a corresponding deletion in the telomeric region of chromosome 8.
3.1. Analysis of Productivity
[0193]The volumetric productivity of C8DEL was evaluated in comparison to the parental cell line from which it was derived. Stable as well as transient transfections were performed.
[0194]Cell cultivation, transfection and screening were carried out in shake flasks using suspension growing CHO cells in a chemically defined culture medium. Cells were transfected by electroporation with different expression vectors encoding a variety of antibodies and therapeutic proteins. Expression vectors used comprised an expression cassette comprising a polynucleotide encoding a neomycin...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com