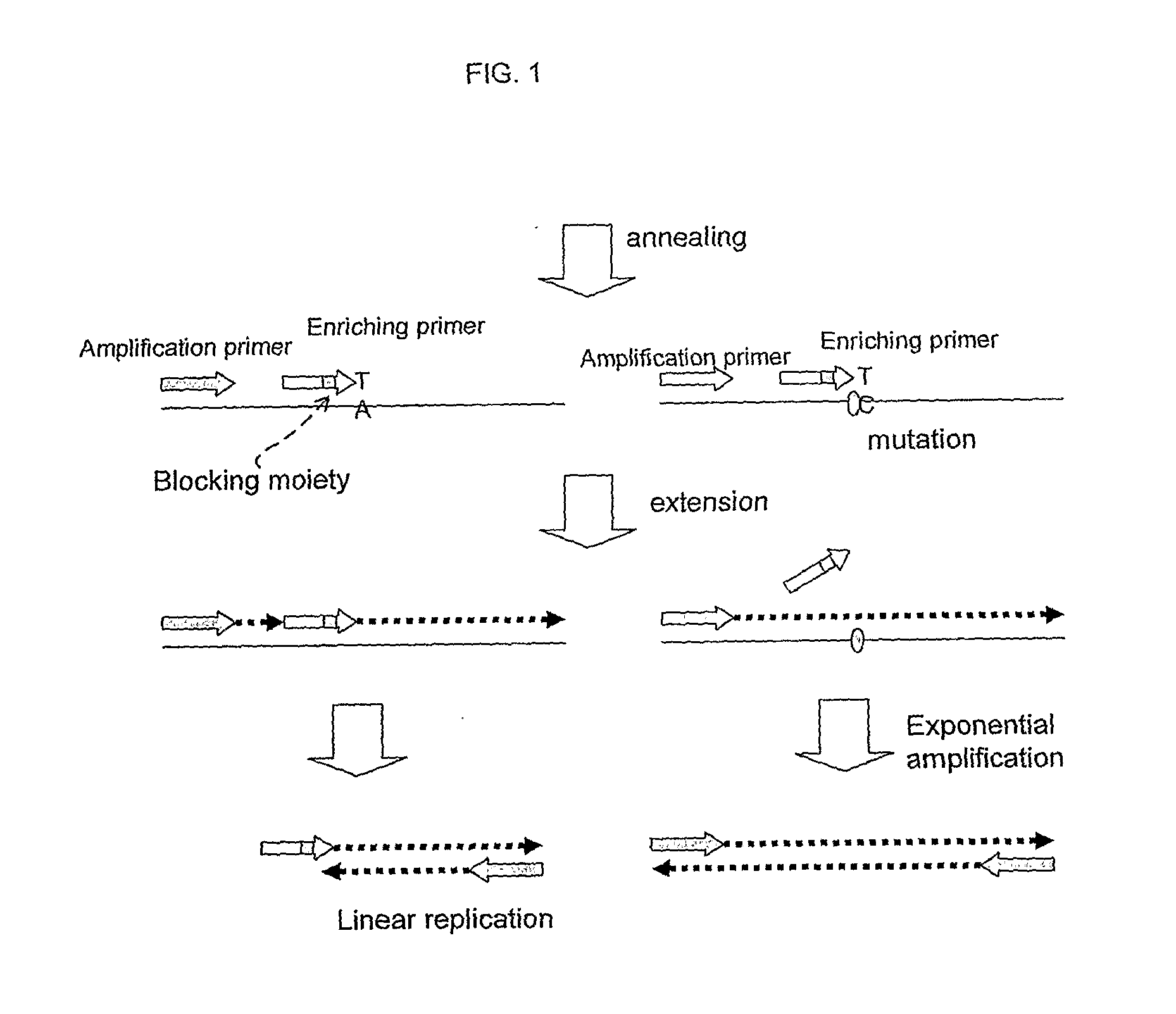
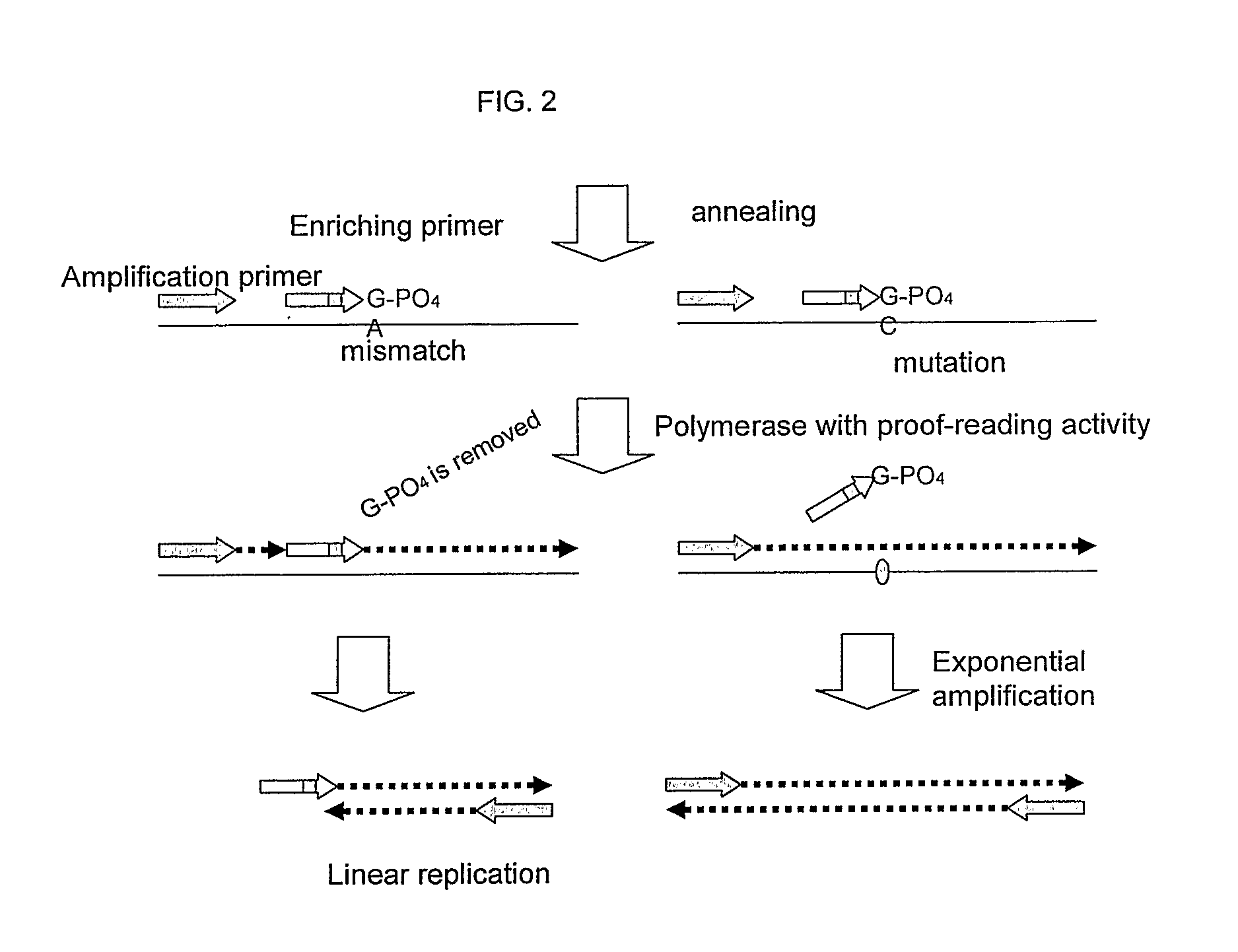
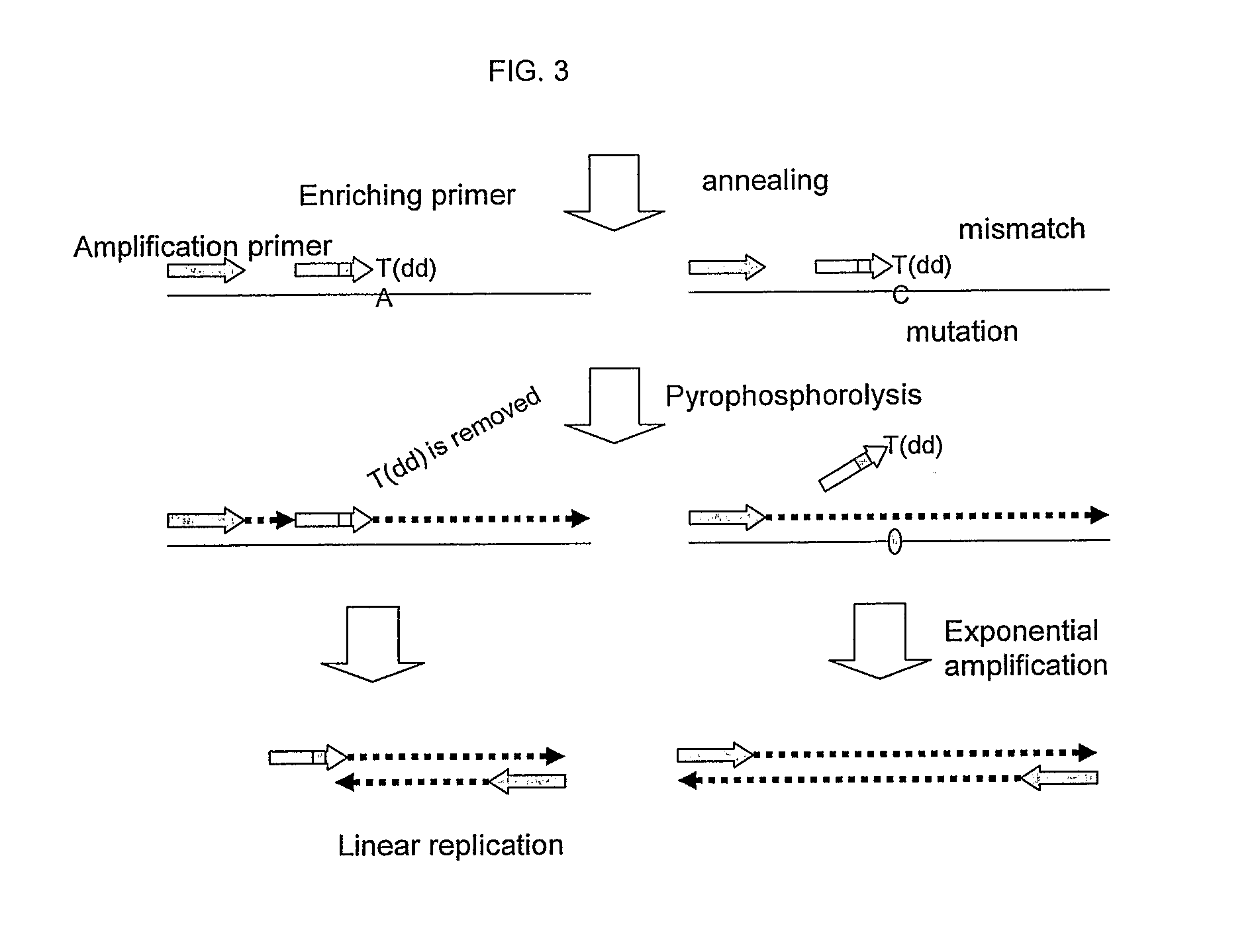
Nucleic acid detection
a nucleic acid and enrichment technology, applied in the field of nucleic acid detection, can solve the problems of false positives, inability to detect single methods, and inability to detect single nucleic acids, etc., to achieve rapid and sensitive detection of genetics, improve the enrichment and detection of desired nucleic acids, and improve the effect of detection speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0148]All primers and probes used in the subsequent experiments were synthesized by EUROGENTEC, UK. Real-time PCR and melting curve analysis were performed on Bio-Rad Chromo4 real-time PCR or Stratagene MX3005P machine. Primers were designed to amplify a target DNA sequence BRAF gene from plasmids comprising a normal BRAF gene fragment and a mutated BRAF gene fragment (harbouring V599E). The sequence of this gene fragment comprises the sequence:
ggaaagcatctcacctcatcctaacacatttcaagccccaaaaatcttaaaagcaggttatataggctaaatagaactaatcattgttttagacatacttattgactctaagaggaaagatgaagtactatgttttaaagaatattatattacagaattatagaaattagatctcttacctaaactcttcataatgcttgctctgataggaaaatgagatctactgttttcctttacttactacacctcagatatatttcttcatgaagacctcacagtaaaaataggtgattttggtctagctacagtgaaatctcgatggagtgggtcccatcagtttgaacagttgtctggatccattttgtggatggtaagaattgaggctatttttccactgattaaatttttggccctgagatgctgctgagttactagaaagtcattgaaggtctcaactatagtattttcatagttcccagtattcac
[0149]The sequences of primers are:
BrafF2GGAAAGCATCTCACCTCATCC...
example 2
[0153]Melting curve analysis of a bridge-probe hybridised to matched and mismatched templates
[0154]A bridge-probe PadLfam is designed having a sequence GAGCCGTCGGTGGTCaaaaaaaaaaCATGACGAGCCCTA, wherein the 5′ end is labelled with 6-Fam, and the 3′ end is labelled with BHQ1. The Binding portions are in uppercase letters, the bridging portion is in lowercase letters.
[0155]Two template oligos are also designed which are: PadLTemp ATAGACCACCGACGGCTCATTAGGGCTCGTCATGTAAC, and PadLTempM ATAGACCACCGACGGCTCATTAGGGCTAGTCATGTAAC, wherein PadLTempM contains a single nucleotide difference which is underlined.
[0156]The bridge-probe PadLfam hybridises to its templates in a form like FIG. 6A. A melting curve analysis was performed as shown in FIG. 9A. A 6 degree Tm difference is observed between matched template (line 2) and template with a single nucleotide difference (line 1) hybridised to the bridge-probe.
example 3
Different Length of the Bridging Portions of a Bridge Probe Affects Tm
[0157]Two bridge-probes SunDab and SunDabA13 are designed as follows, wherein the 3′ ends are labelled with Dabcyl. The probe SunDab has no bridging portion, whereas probe SunDabA13 contains a bridging portion with 13 As. A second probe SunFam contains two regions complementary to the first binding portion and second binding portion of the bridge-probe and is labelled with 6-Fam at 5′ end. Another oligonucleotide SunTemp is designed to have a region complementary to a part of the second probe. Alignments showing the binding regions are as follows, wherein “F” means 6-Fam, “Q” means dabcyl, “−” means no nucleotide, “I” means complementary bases, “P” indicates phosphate group.
SunFam5′ FCACCGCGCTTAGTTACATGACGAGCCGTGTAGCGTGGACGACAGAGG-P 3′ IIIIII IIIIIIIIIIIIIIIIIIIIISundab3′ QGTGGCG--------------------CACATCGCACCTGCTGTCTCC 5′SunFam5′ FCACCGCGCTTAGTTACATGACGAGCCGTGTAGCGTGGACGACAGAGG-P 3′ IIIII...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Concentration | aaaaa | aaaaa |
| Electrical resistance | aaaaa | aaaaa |
| Melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com