Integrated Analyses of Breast and Colorectal Cancers
a technology of integrated analysis and breast cancer, which is applied in the field of breast and colorectal cancer, can solve the problems of difficult detection of primary tumors, infrequent detection of high copy amplification or homozygous deletion (hds), and difficult identification of focal hds
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
Optimization of Copy Number Analysis with Digital Karyotyping
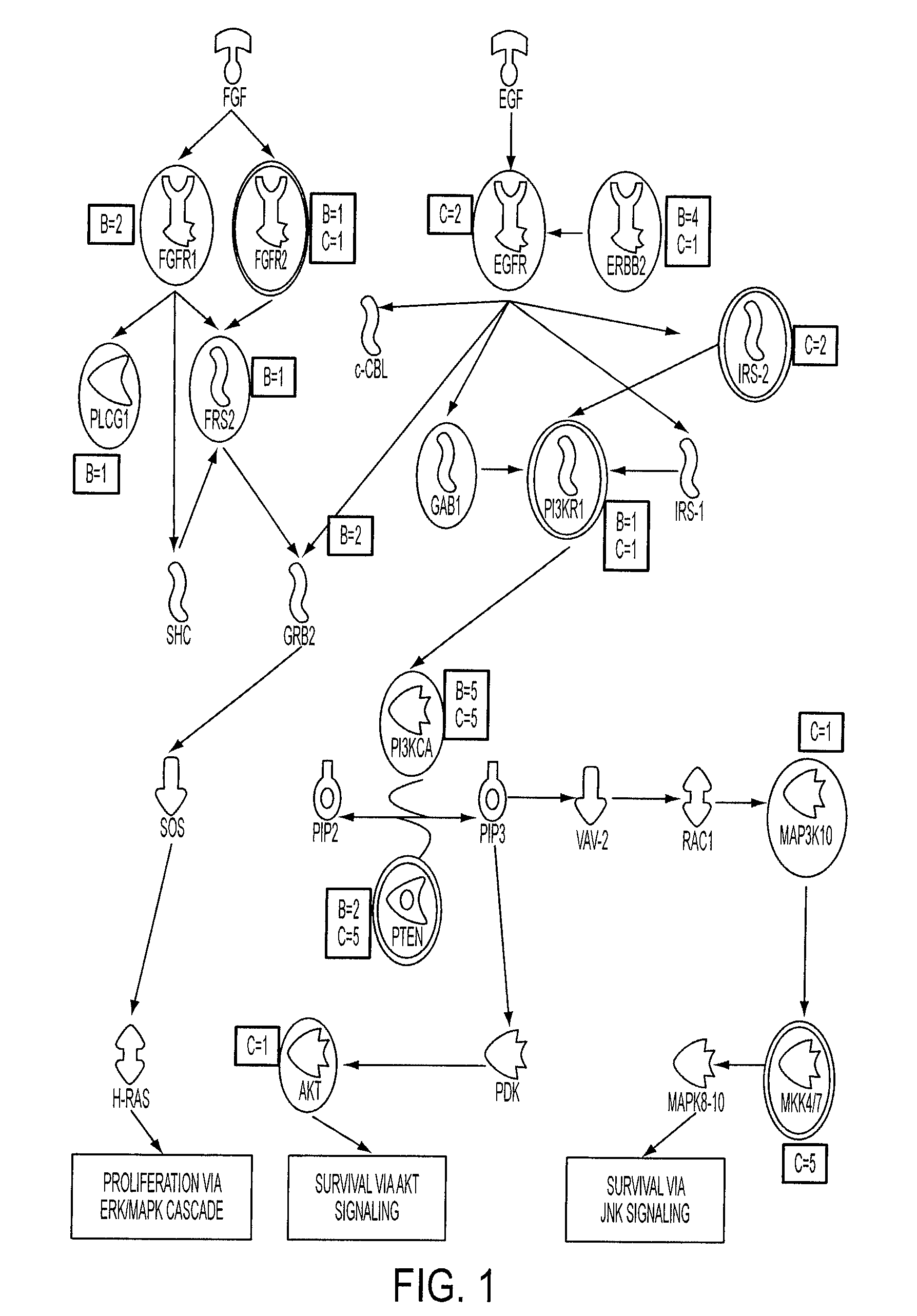
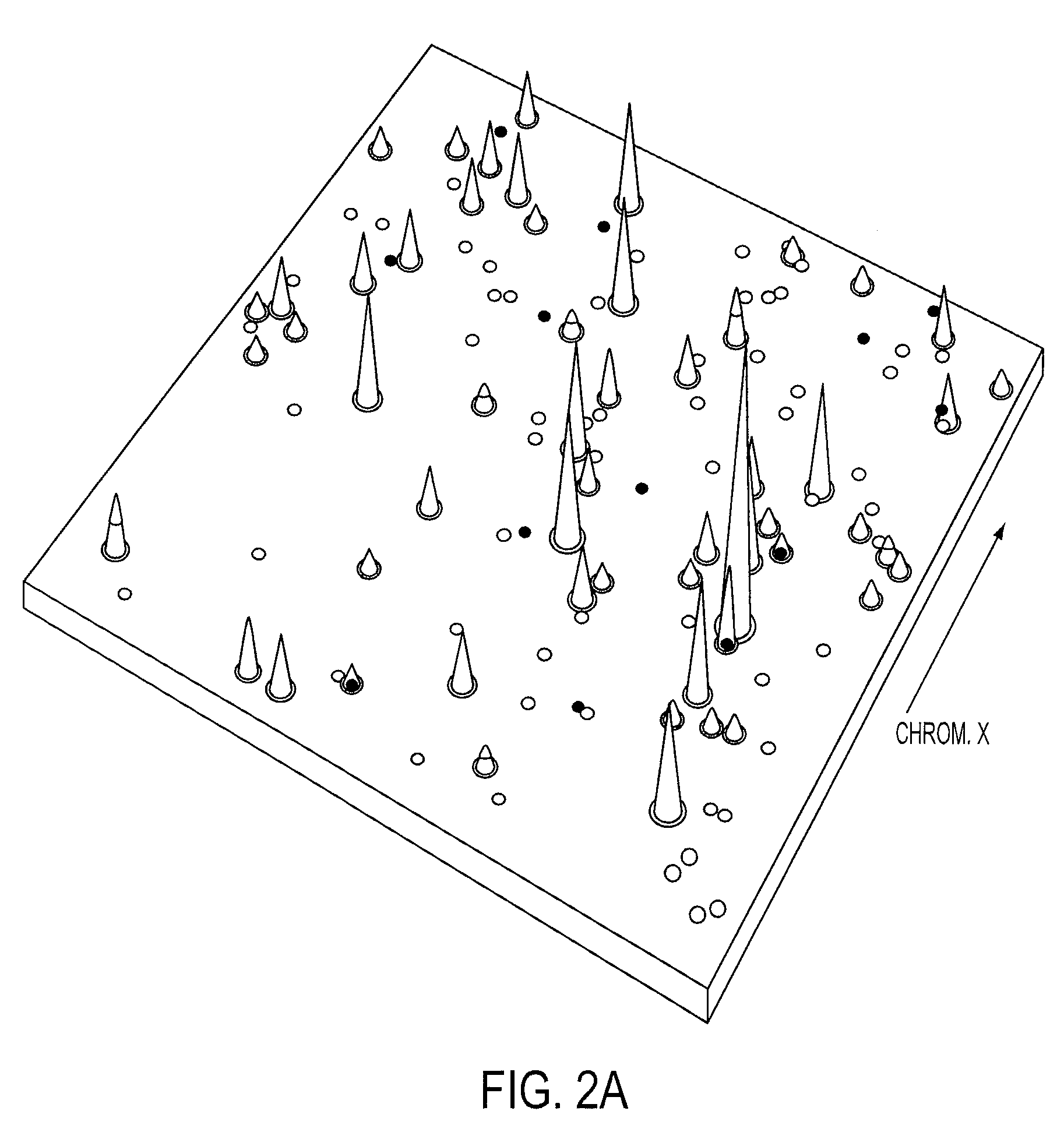
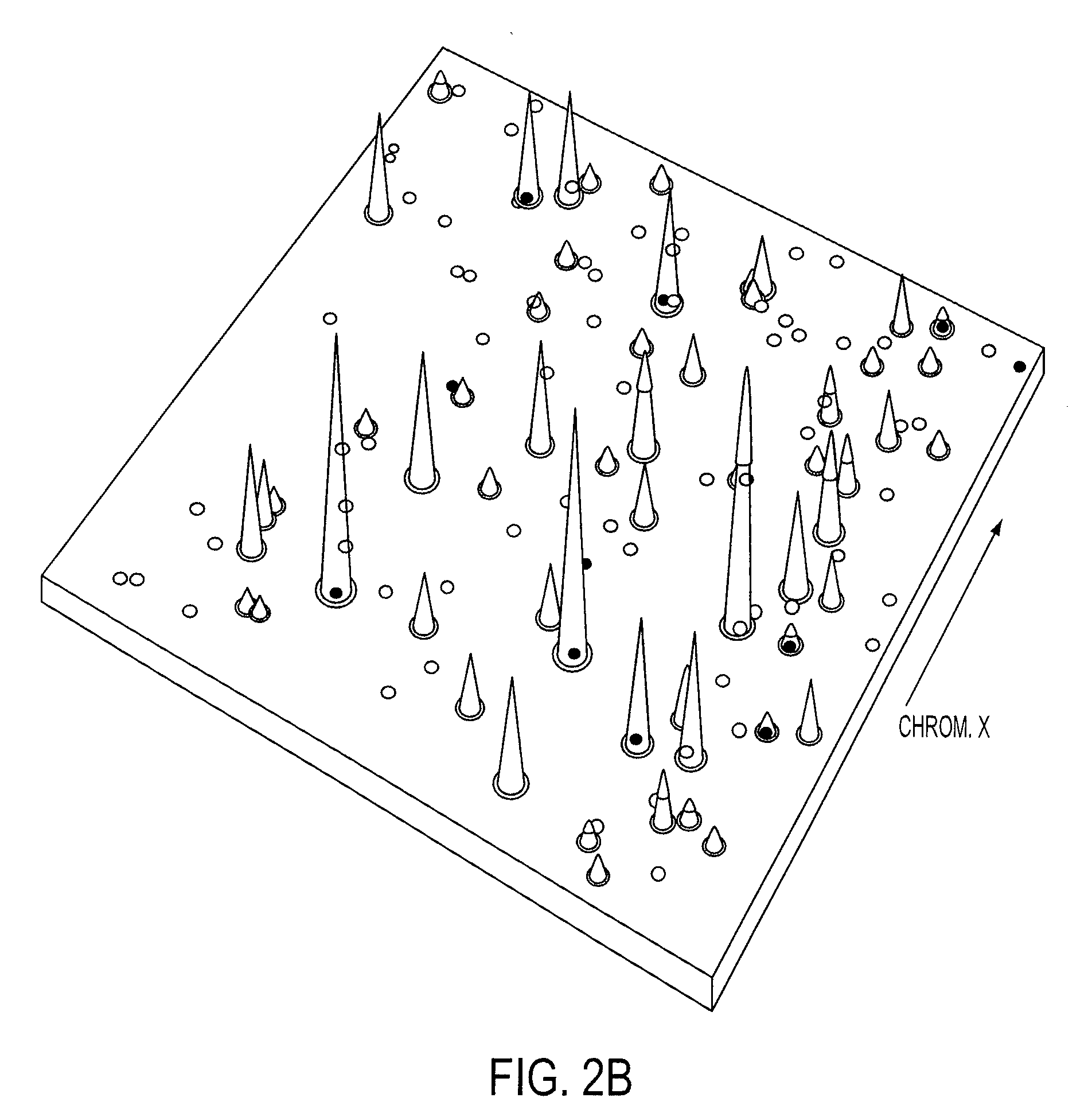
[0042]Digital Karyotyping (DK) was used as a standard to develop criteria for assessing amplifications and HDs with Illumina high density SNP arrays. Analysis of DK libraries from 18 colorectal tumor samples identified a total of 21 amplification events, each containing relatively small chromosomal regions (41 kb to 2.3 Mb) with 12 to 186 copies per nucleus (SI Table 2). We also found 4 regions within the autosomal chromosomes where the tag density reached zero, representing HDs. As expected, we identified low-amplitude gains and losses of entire chromosomes, chromosomal arms, or other large genomic regions. We did not pursue these low-amplitude copy number changes as it is difficult to reliably identify candidate cancer genes from such large regions. To ensure that the copy number changes identified by DK were bona fide amplifications or HDs, we independently examined 12 alterations by quantitative PCR and confirmed the p...
example 2
Detection of Amplifications and Homozygous Deletions
[0045]A total of 45 breast and 36 colorectal tumors were analyzed by Illumina arrays containing either ˜317,000 or ˜550,000 SNPs (SI FIG. 1). To determine the fraction of alterations that were likely to be somatic (i.e., tumor derived), we analyzed these regions in 23 matched normal samples. In the normal samples, no amplifications and only four distinct HDs were detected. We removed these alterations from further analysis, as well as those corresponding to known copy number variation in normal human cells (27, 28). Finally, we removed any copy number changes where the boundaries were identical in two or more samples, as these were likely to represent germline variants. Based on this conservative strategy, we estimated that >95% of the 614 amplifications and 463 HDs (SI Table 3) represented true somatic alterations.
[0046]Breast cancers contributed to a majority of the alterations identified, comprising 68% and 81% of the total HDs ...
example 3
Genes Altered in More than One Tumor
[0048]One of the main challenges in the analysis of somatic alterations in cancers involves the distinction between those changes which are selected for during tumorigenesis (driver alterations) from those that provide no selective advantage (passenger alterations). Even in regions that have multiple copy number alterations, this distinction can be particularly difficult because regions of amplification and HD can contain multiple genes, only a subset of which are presumably the underlying targets. We reasoned that the integration of copy number analyses with sequence data would help reveal the driver genes that were more likely to contain genetic alterations. To accomplish this integration, we developed a new statistical approach for determining whether the observed genetic alterations of any type in any gene were likely to reflect an underlying mutation frequency that was significantly higher than the passenger rate. To analyze the probability t...
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
| density | aaaaa | aaaaa |
| purity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com