Long distance polymerase chain reaction-based assay for detecting chromosomal rearrangements
a polymerase chain reaction and assay technology, applied in the field of coagulation, can solve the problems of common unamplification, and difficult optimization of primer concentration
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Sequence for Flanking Regions of Factor VIII Gene Int22h1, Int22h2 and Int22h3
[0059]Fifty bp of 5′ flanking sequence of Int22h1 in intron 22 was available from GenBank (Accession Nos. X86011 and X86012). Fifty bp of flanking sequences were also known at both 5′ and 3′ ends of Int22h2 and Int22h3 (Naylor et al., 1995). Inverse-PCR (Ochman et al., 1988) was applied to amplify the unknown flanking sequences.
[0060]To obtain sequence for 5′ flanking Int22h1, a 6 kb segment was amplified with the Expand™ Long Template PCR system (Boehringer Mannheim). The primers used were: F8(E22)(89)32D: 5′-TGCCCGTCAGAAGTTCTCCAGCCTCTACATCT-3′ (SEQ ID NO:1) (bases 89-120 of Genbank Accession No. M88644) which is a primer corresponding to exon 22 of the factor VIII gene and Int22h(365)32U: 5′-GGTCAAGACTGAAATTAGCGTGTTAGGCAAGA-3′ (SEQ ID NO:2) (bases 384-415 of GenBank Accession No. X86011) which corresponds to the 5′ region of Int22h1. An ABI Model 377 sequencer was utilized to obtain 1319 bp of the 5′ fla...
example 2
Primer Design for the PCR Assay
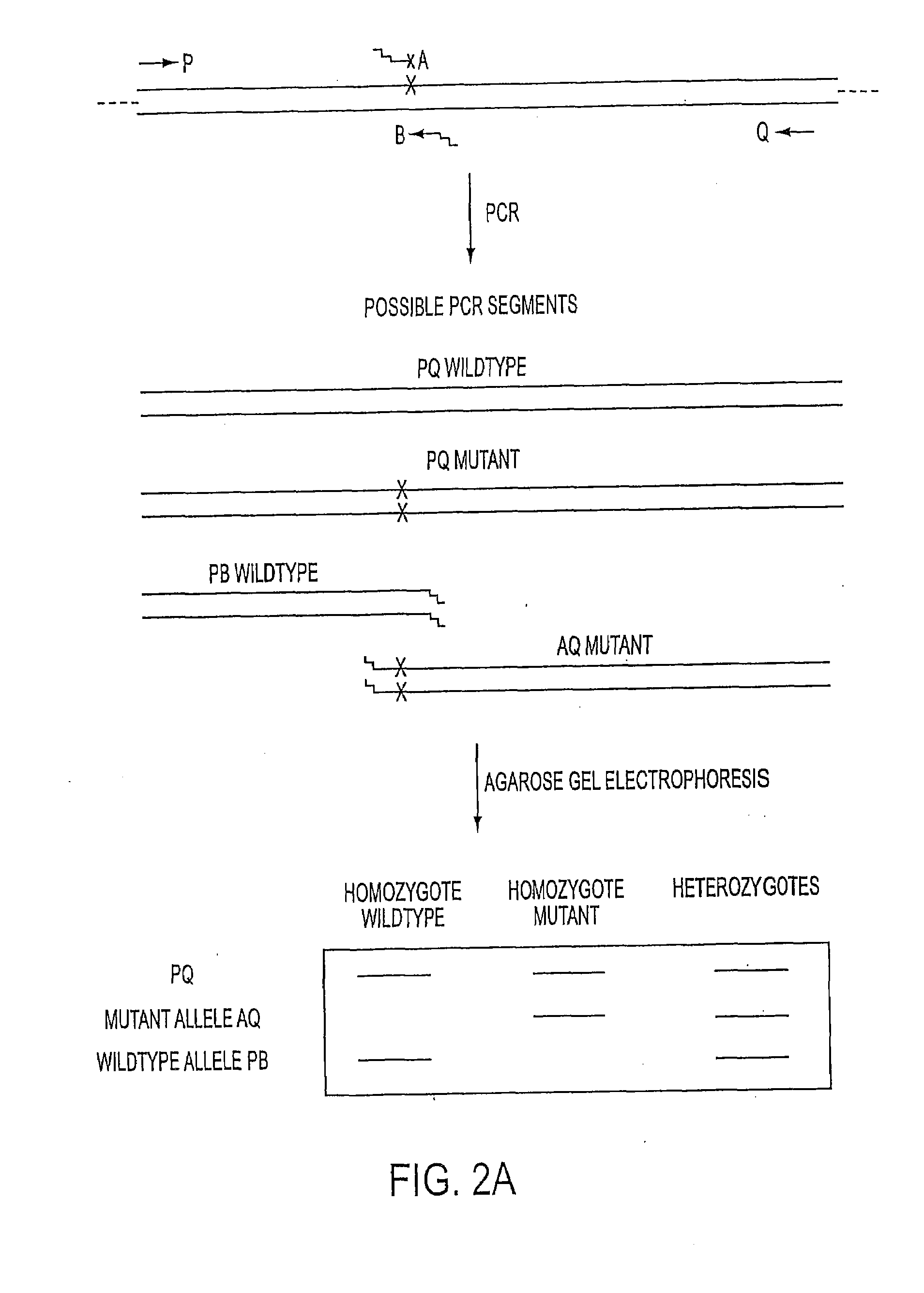
[0063]Four primers are used in a multiplex PCR assay to detect the inversion in the factor VIII gene. These primers are labeled P, Q, A and B and their locations are indicated in FIG. 3A. These primers were designed with the aid of Oligo 5 software (National Biosciences). The Tm value of each PCR segment was estimated by the formula of Wetmur: Tmproduct=81.5+16.6 log [K+]+0.41% (G+C)−675 / length (Wetmur, 1991). The G+C content of the AB segment is 51% on average and the Tm is 81° C. The Tm values of the primers were estimated by the nearest neighbor method at 50 mM KCl and 250 μM DNA with the formula: Tmprimer=ΔH / {ΔS+R×ln(C / 4)}+16.6 log [K+]−273.15 (Breslauer et al., 1986; Freier et al., 1986). Each primer was designed to have a Tm that was 10° C. lower than the average Tm of the PCR products (73° C.). Primer lengths varied from 36-40 nucleotides. For primers P, Q and B, a high GC tail of two to five nucleotides was added in order to achieve Tm value of...
example 3
The PCR Assay
[0065]The PCR was performed from human genomic DNA isolated from white blood cells, although DNA from other cells could also be used. Unless otherwise stated, the PCR mixtures contained a total volume of 25 μL: 50 mM Tris-HCl, pH 9.2, 2.25 mM MgCl2, 16 mM (NH4)2SO4, 7.5% DMSO, 500 μM of dGTP and deaza-dGTP (62.5%:37.5% or 50%:50%), 500 μM of each of the other dNTPs, 250 ng of genomic DNA.
[0066]Three types of cycling conditions were utilized: three-temperature PCR, two-temperature PCR and S-PCR. The cycling conditions for three-temperature PCR were 94° C. for 12 seconds, 65° C. for 30 seconds and 68° C. for 14 minutes for the first 10 cycles (Perkin Elmer GeneAmp PCR System 9600). The remaining 20 cycles were performed by adding an extra 20 seconds to the elongation per cycle.
[0067]Conditions for two temperature cycling were: 94° C. for 12 seconds and 65° C. for 15 minutes for the first 10 cycles, with an extra 20 seconds added to the elongation per cycle for the remaini...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com