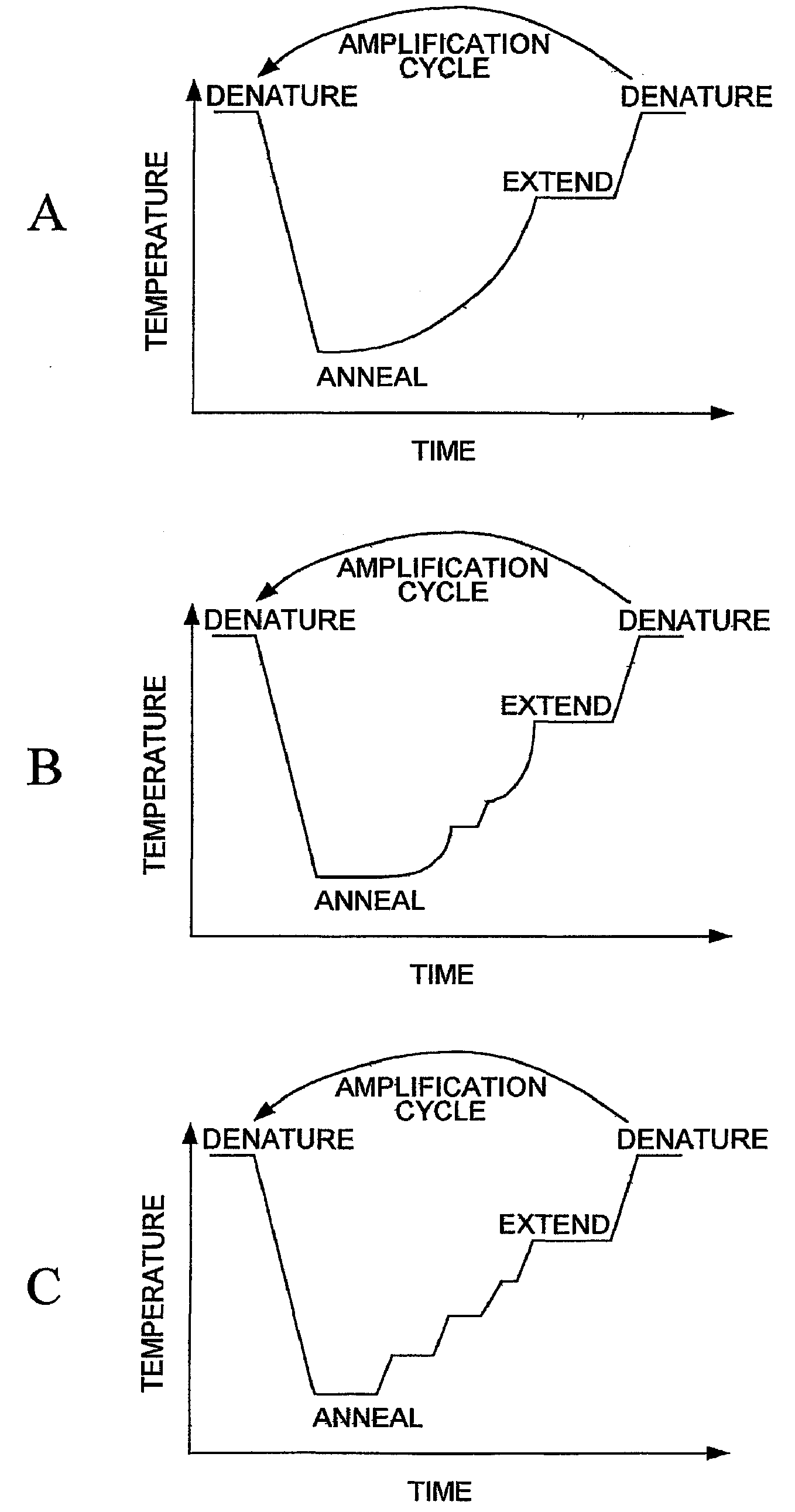
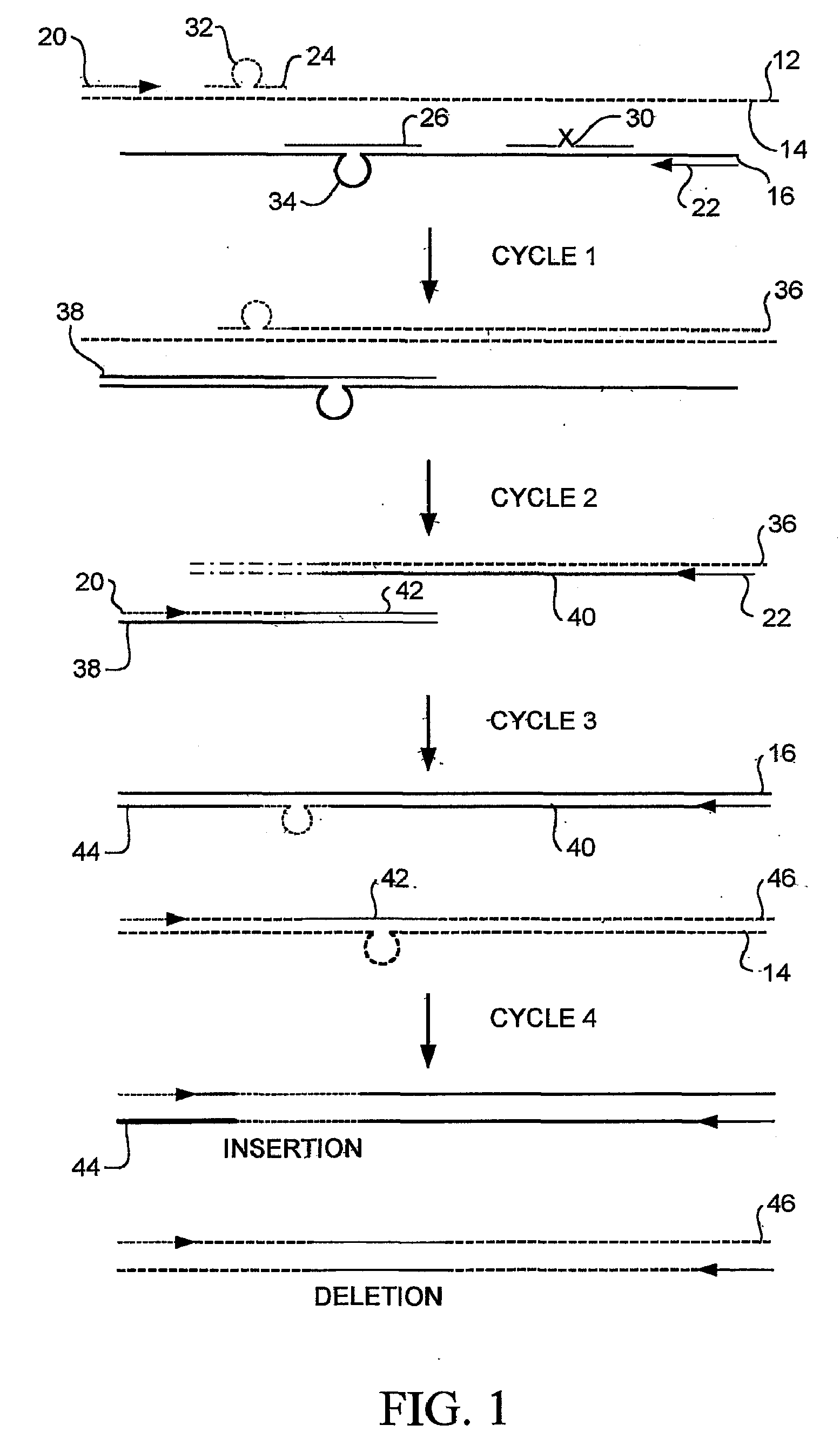
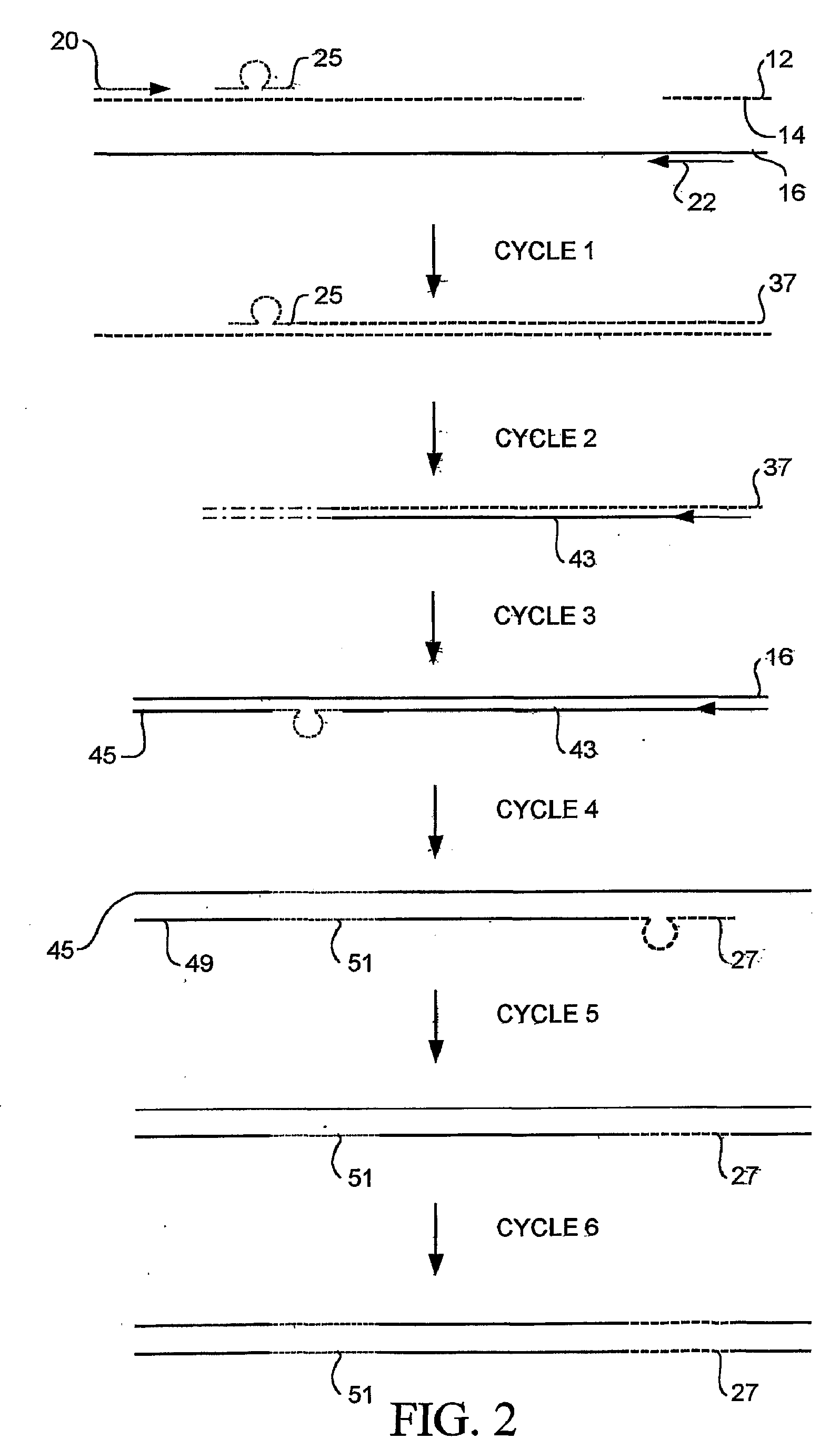
Random Mutagenesis And Amplification Of Nucleic Acid
a nucleic acid and mutagenesis technology, applied in combinational chemistry, dna preparation, library creation, etc., can solve the problems of tertiary structure, biological function remains elusive, and formidable tasks, and achieve the goal of promoting heterologous binding of oligonucleotides, enhancing degeneracy of oligonucleotides, and increasing the efficiency of extension of oligonucleotides
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example
[0166]The gene encoding a penicillinase from Bacillus licheniformis was used as a target to be randomly mutagenized. By randomly mutating the enzyme, isozymes which show altered hydrolytic activity and / or specificity against various penicillins and cephalosporins may offer clues to 1) how antibiotics can be designed to thwart the inevitable evolution towards β-lactamases which render pathogenic bacteria resistant to drug therapy, and 2) offer further information for the study of protein structure-function relationships.
[0167]The gene encoding the Bacillus licheniformis was isolated from a plasmid pELB1. The plasmid pELB1 is a pBR322 derivative, containing the “exolarge” form of the B. licheniformis b-lactamase gene, utilizing the Bacillus amyloliquefaciens promoter and subtilisin signal sequence, and Bacillus and E. coli origins of replication (Ellerby, L. M., Escobar, W. A., Fink, A. L., Mitchinson C., Wells J A (1990) Biochemistry, June 19; 29(24):5797-806).
[0168]pELB1 was digeste...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Time | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com