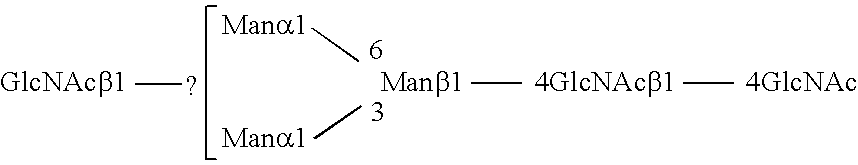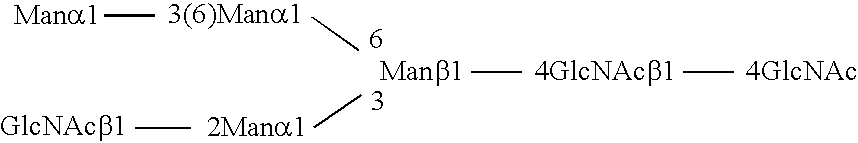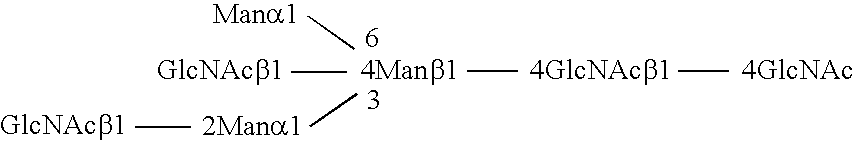Methods and products related to the improved analysis of carbohydrates
a technology of improved analysis and carbohydrates, applied in the field of improved analysis of carbohydrates, can solve the problems of embryonic lethality and enormous multisystemi
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
N-Glycan Analysis
Materials and Methods
PNGaseF Digest of N-Glycans from Protein Cores
[0219] Between 10 and 100 μg of protein were denatured for 10 minutes at 90° C. with 0.5% SDS and 1% β-mercaptoethanol. Since SDS (and other ionic detergents) inhibits enzyme activity, 1% NP-40 was added to counteract these effects. The enzyme reaction was performed overnight with 2 μl of PNGaseF at 37° C. in a 50mM sodium phosphate buffer, pH 7.5.
Purification of Released N-Glycans
[0220] Proteins were precipitated with a 3× volume of 100% ethanol on ice for 1 hour. After centrifugation to remove the proteins, the supernatant containing the N-glycans was evaporated by vacuum (SpeedVac, TeleChem International, Inc., Sunnyvale, Calif.). Dried glycans were resuspended in 50 μl of water.
[0221] Samples were desalted using 1 ml ion exchange column of AG50W X-8 beads (Bio-Rad, Hercules, Calif.). The resin was charged with 150 mM acetic acid and washed with water. Glycan samples were loaded onto the ...
example 2
Profiling of N-Glycans from Human Serum
Materials and Methods
[0246] Cleavage of N-glycans from Serum Glycoproteins (Reduction / Carboxymethylation Method)
[0247] Human male normal serum samples were obtained from IMPATH (Franklin, Mass.) and Biomedical Resources (Hatboro, Pa.), and stored at −85° C. For each experiment, 50 μl of serum was used to harvest N-glycans. Serum samples were first diluted 1:4 with water, then DTT was added to a final concentration of 80 mM. After incubation for 30 minutes at 37° C., iodoacetic acid was added to a final concentration of 400 mM and incubated for 1 hour more at 37° C. The sample was dialyzed against 10 mM Tris acetate pH 8.3 overnight and concentrated to ˜200 μl in a spin column with a 3000 Da Molecular Weight Cut off (MWCO) filter (VivaScience, Hannover, Germany). To cleave the sugars from the protein, 5 μl (1,000 U) of PNGaseF (New England Biolabs, Beverly, Mass.) was added and allowed to react overnight at 37° C.
Purification of N-Glycans ...
example 3
Glycan Analysis
Release of Glycans from Proteins
[0284] Several methods were used to cleave the carbohydrates from proteins:
[0285]
[0286] A) Glycoproteins were denatured with 0.5% SDS and 1% β-mercaptoethanol. Since SDS (and other ionic detergents) inhibits enzyme activity, 1% NP-40 was added to counteract these effects. The enzymatic cleavage was performed overnight with PNGaseF (New England Biolabs) at 37° C. in sodium phosphate buffer, pH 7.5 or Tris acetate buffer pH 8.3.
[0287] B) Samples were reduced with DTT followed by alkylation with either iodoacetic acid or iodoacetamide. The sample was dialyzed against phosphate buffer, pH 7.5 or Tris acetate, pH 8.3 overnight and concentrated to ˜200 μl in a spin column with a 3000 Da MWCO filter. To cleave the sugars from the protein between 100 and 2,000 U of PNGaseF (New England Biolabs) were used.
[0288] C) Glycoproteins were denatured using a buffer containing 8M urea, 3.2 mM EDTA and 360 mM Tris, pH 8.6 (Papac, D. I., et al. 1998...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com