Methods and compositions for tailing and amplifying RNA
a technology of rna and rna molecules, applied in the field of molecular biology, can solve the problems of ineffective capture of rna molecules lacking poly(a) tails, inability to fully realize in vivo whole genome expression analysis methods, and ineffective capture methods of mrna purification from eukaryotic cells, etc., to achieve efficient and uniform tailing of targeted rna
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Optimized Polyadenylation of Bacterial mRNA
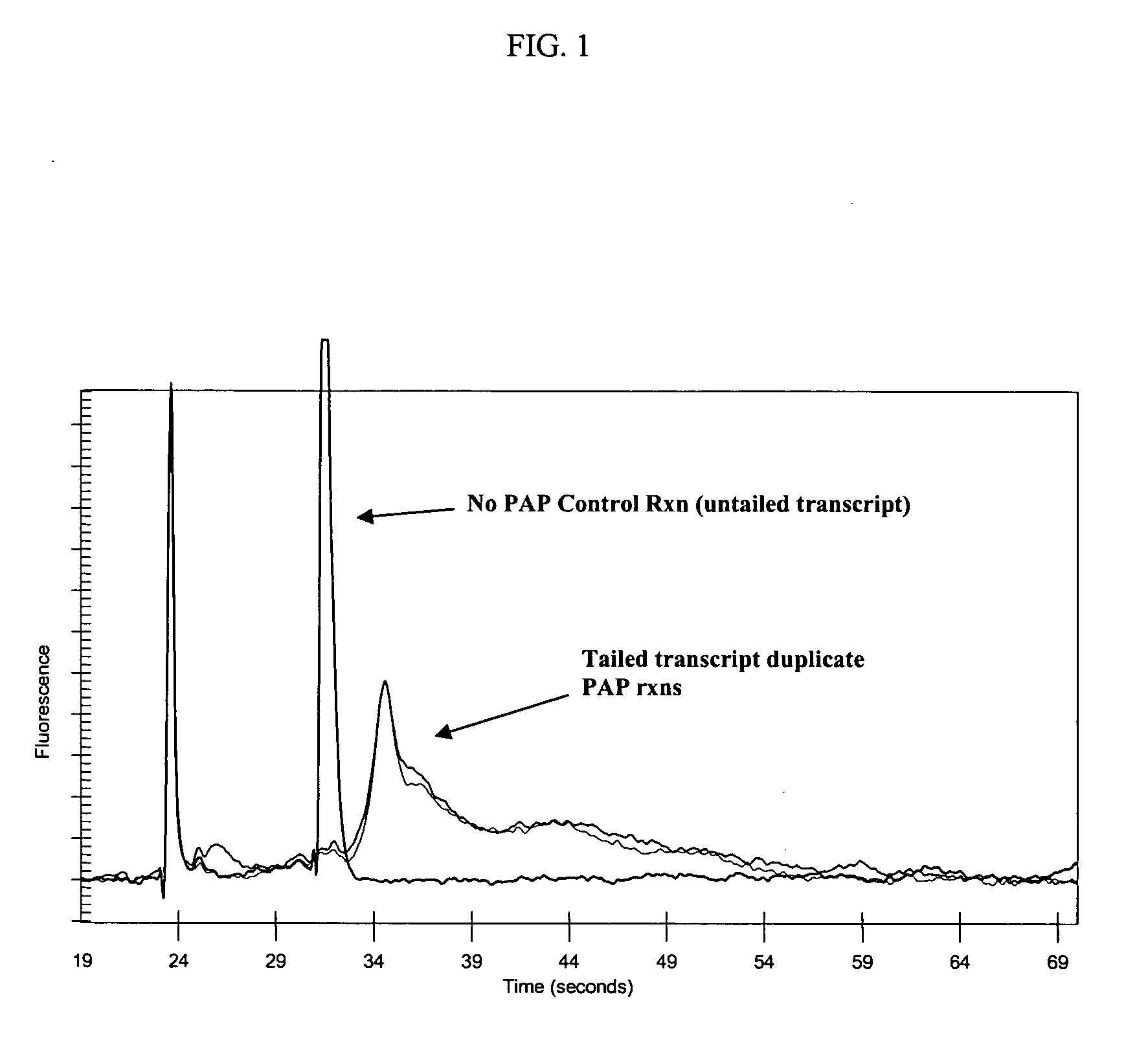
[0145] Seemingly straightforward and simple, the polyadenylation of complex RNA populations is actually a quite complicated reaction. For polyadenylating bacterial mRNAs, the inventors used purified poly(A) polymerase I (PAP I) enzyme of E. coli. PAP I or PAP adds AMP to 3′ ends of RNA molecules using ATP as a donor. The efficiency of PAP has been found to vary with different substrate RNAs. RNAs with 3′ end nucleotides that are paired, such as in a hairpin, are less efficiently polyadenylated (Feng and Cohen, 2000). The enzyme uses an ordered bi-bi kinetic mechanism and adds adenines distributively (Eun, 1996). Initiation and elongation occur at different rates, and the enzyme dissociates from the primer after every step.
[0146] The initial goal was to ensure that all mRNAs in a population of bacterial mRNA were poly(A) tailed, thereby ensuring that they will be templates for the 1st strand cDNA synthesis reaction—the first step in amplif...
example 2
Effect of MnCl2 Concentration on Polyadenylation Reaction
[0153] This experiment was designed to test whether or not the addition of MnCl2 has an effect on poly(A) tailing efficiency and tail length. The reaction utilized 100 ng of a synthetic in vitro transcribed RNA of 387 nucleotides as template. The reactions were performed as follows:
[0154] RNA template (100 ng) and H2O to a final volume of 10 μl were combined in a 0.5 ml tube. The RNA solution was heat denatured at 70° C. for 10 minutes and then quenched on ice for 3 minutes. The other reactants were then added as a 15 μl cocktail containing: 5 μl 5× Polyadenylation buffer, 1.25 μl 1 mM ATP, 1 μl Ribonuclease inhibitor (40 U / μl), 1 μl E. coli Poly(A) Polymerase (2 U / μl) and 4.25 μl H2O. Depending on the reaction (with MnCl2 or without MnCl2) either 2.5 μl 25 mM MnCl2 or 2.5 μl H2O was also added. The reactions were allowed to incubate at 37° C. for various time points. At the end of this incubation 2 μl of 0.5 M EDTA was adde...
example 3
Bacterial RNA Amplification
[0156] In this experiment total E. coli RNA was polyadenylated and amplified. Two RNA preparations were used (lot # PR043921 and the same RNA after purification over a glass fiber filter column) for amplification and two in vitro transcription (IVT) incubation times were compared (6 hours and 14 hours).
[0157] Total RNA template (10 ng or 100 ng) and H2O to a final volume of 5 μl were combined in a 0.5 ml tube. The RNA solution was heat denatured at 70° C. for 10 minutes and then quenched on ice for at 3 minutes. The other reactants were then added as a 5 μl cocktail containing: 1× Polyadenylation buffer (50 mM Tris-HCl pH 8.3, 75 mM KCl, 3 mM MgCl2 and 10 mM DTT), 50 μM ATP, 40 Units Ribonuclease inhibitor Protein, 2 Units E. coli Poly(A) Polymerase. The reaction was incubated for 15 minutes at 37° C. and placed on ice until the amplification step.
[0158] Ambion's MessageAmp II RNA amplification kit was used to amplify the polyadenylated bacterial RNA ac...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com