Oligonucleotide sizing using cleavable primers
a technology of oligonucleotide and primer, which is applied in the field ofoligonucleotide compositions containing cleavable primers, can solve the problems of reducing so as to achieve the effect of increasing the amount of new or useful fragment information for primer extension products, improving the present invention, and increasing the number of fragments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Preparation of a Modified Oligonucleotide Containing A 3′-5′-Cleavable Linkage
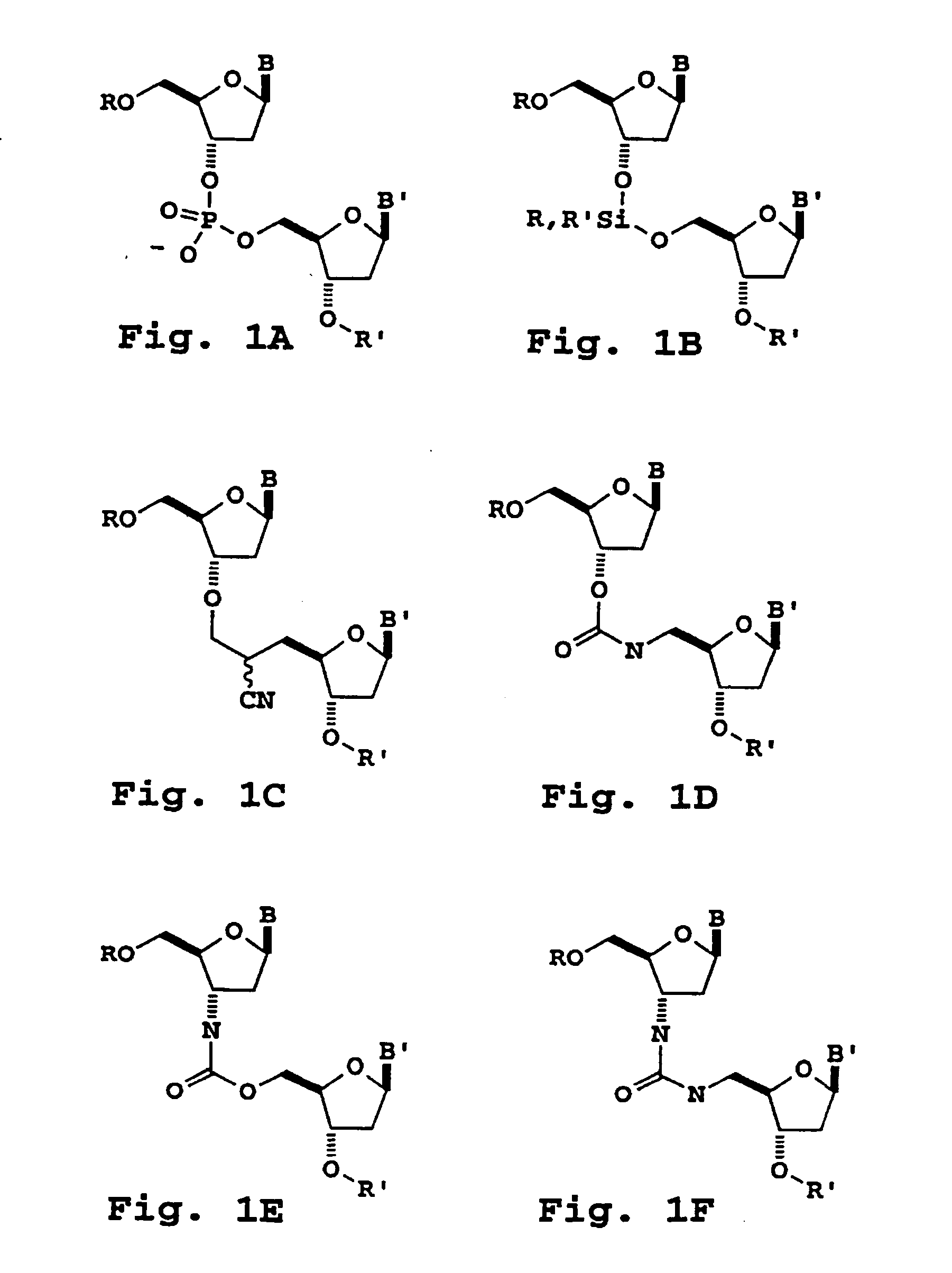
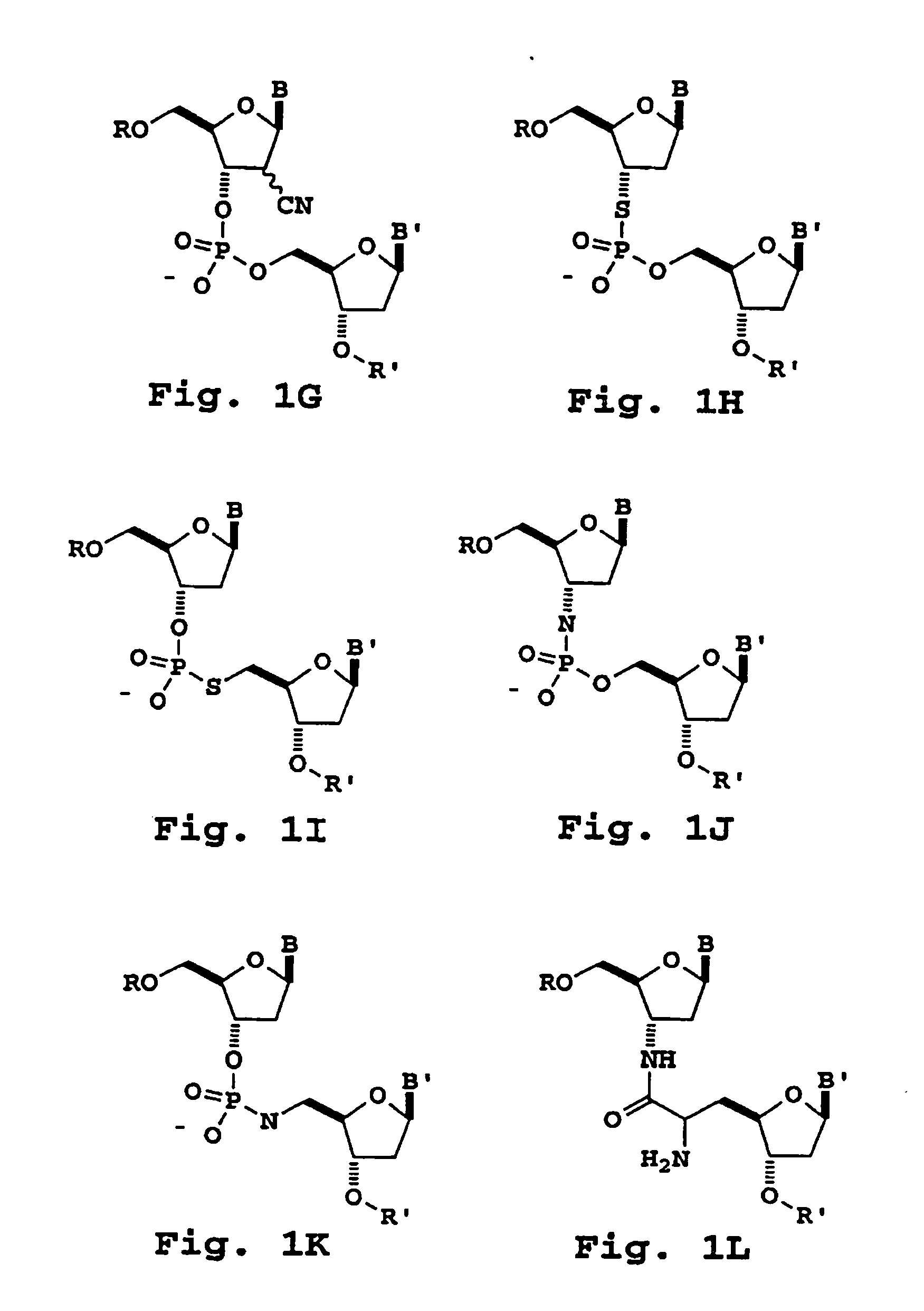
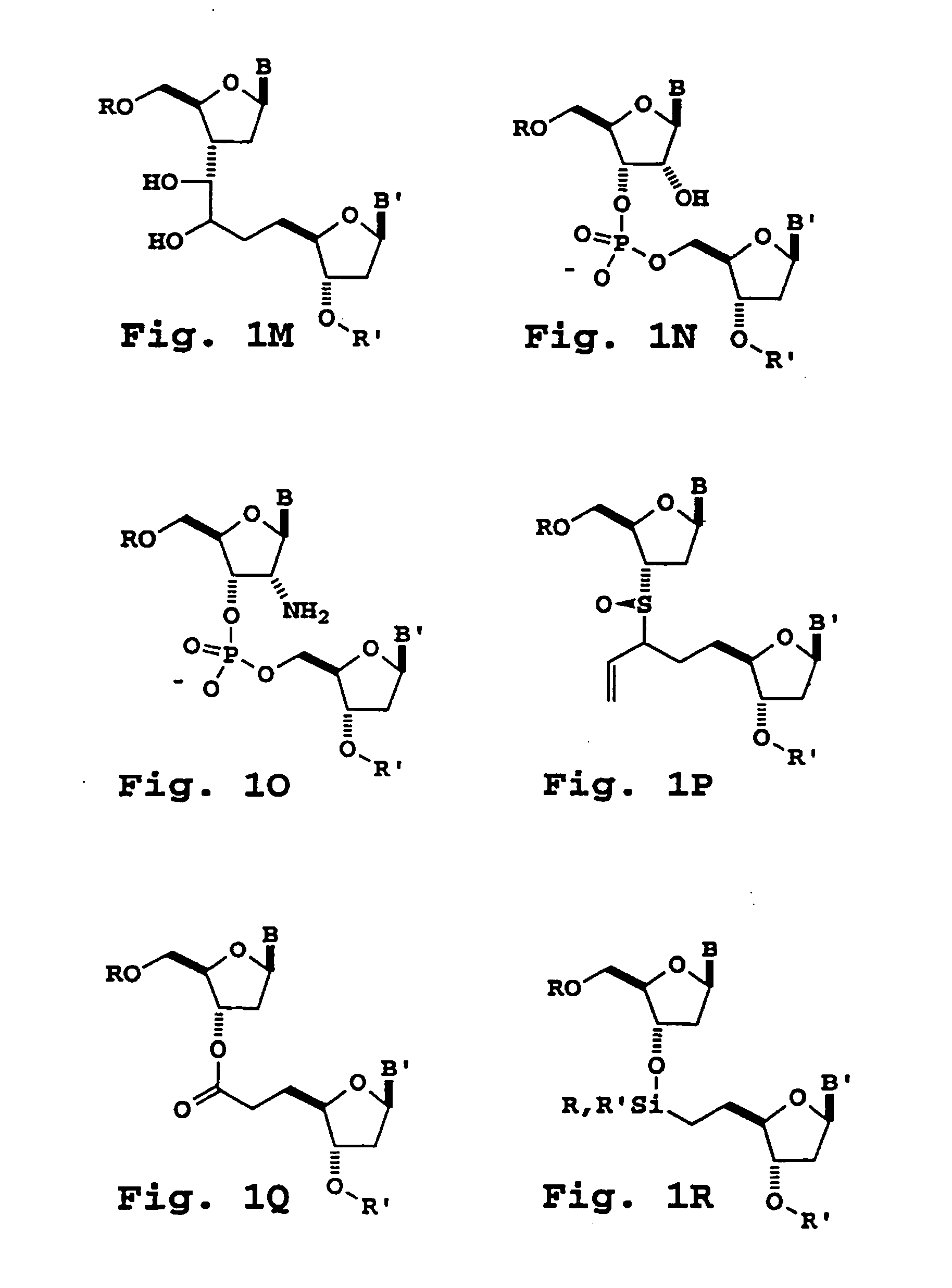
[0292] Nucleoside dimers containing the following 3′-5′-internucleoside cleavable linkages are prepared as follows.
A. 3′,5′-Dialkoxysilane Internucleoside Linkage
[0293] The 3′-O-functionalized nucleoside intermediate, 3′-O-(diisopropylsilyl)-2′-deoxynucleoside triflate (1) is prepared by first adding bis(trifluoromethanesulfonyl)diisopropylsilane (1 mmol) to an equimolar amount of the sterically hindered base, 2,6-di-tert-butyl-4-methylpyridine, dissolved in dry acetonitrile, under an inert atmosphere. The resulting solution is cooled to −40° C. in a cooling bath, to which is added a solution of a 5′-(O)-protected nucleoside, 5′-(dimethoxytrityl)-2′-deoxynucleoside (0.9 mmol) and 2,6-di-tert-butyl-4-methylpyridine (0.2 mmol) in dimethylformamide over a 10 min period. The resulting reaction mixture is stored at 40° C. for 1 h and then allowed to warm to room temperature. The 3′-O-diisopropylsilyl triflat...
example 2
Attachment to the Solid Support
A. Streptavidin Affinity Immobilization
[0318] A modified primer from Example 1 above containing a cleavable site is immobilized by attachment to a functionalized solid support material. In some cases the cleavable site-containing primer is modified as described below.
[0319] For attaching an oligonucleotide primer to a streptavidin-coated support a biotinylated primer is typically used. Biotinylation is carried out as follows.
[0320] A primer containing a cleavable site is prepared as in Example 1, with a minor modification: the primer is synthesized to contain a reactive amino site for biotinylation. An amino group is introduced during solid phase synthesis using a standard DNA synthesizer, such as Applied Biosystems 393 DNA / RNA Synthesizer.
[0321] To selectively introduce the internal amino function, the modified nucleoside phosphoramidite Amino-Modifier dT, containing a base labile trifluoroacetyl group protecting a primary amine attached to thymi...
example 3
Selective Cleavage of a Synthetic DNA Probe Containing: Cleavable Ribose
[0331] A synthetic DNA probe containing a cleavable ribose site was selectively cleaved by ammonium hydroxide treatment. The 17-mer, having the sequence: 5′-AAA TAC ATC riboGCT TGA AC-3′ (SEQ ID NO:10) was prepared to contain a cleavable ribose in the 7-position. The modified probe was treated with aqueous 3% ammonium hydroxide for 15 min at room temperature (pH 10) to effect selective cleavage of the ribose moiety.
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com