Pigment epithelium-derived factor: characterization, genomic organization and sequence of the PEDF gene
a pigment epithelium and gene technology, applied in the field of neuronotrophic, neuronotrophic and gliastatic proteins, can solve the problem of difficult at best for further study of ped
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0076] This example describes the trypsin digestion of PEDF and the amino acid sequencing of the resulting fragments.
[0077] PEDF was purified from the medium of a primary culture of human fetal RPE cells by high performance liquid chromatography (HPLC). The HPLC-purified PEDF was then reduced and alkylated. Afterwards, it was dried and redissolved in 50 μl of CRA buffer (8 M urea, 0.4 M ammonium carbonate, pH 8.0), and 5 μl of 45 mM dithiothreitol (DTT) (Calbiochem, San Diego, Calif.) were added. After heating at 50° C. for 15 minutes, the solution was cooled, and 5 μl of 100 mM iodoacetic acid (Sigma Chem. Co., St. Louis, Mo.) were added. After 15 minutes, the solution was diluted to a concentration of 2 M urea and subjected to trypsin digestion (Boehringer-Mannheim, Indianapolis, Ind.) for 22 hours at 37° C. using an enzyme:substrate ratio of 1:25 (wt / wt). Tryptic peptides were separated by narrowbore, reverse-phase HPLC on a Hewlett-Packard 1090 HPLC, equipped with a 1040 diode ...
example 2
[0079] This example describes the construction of oligonucleotides, based on the peptide sequences of Example 1, the use of the oligonucleotides in the isolation of PEDF cDNA, and the sequencing of PEDF cDNA.
[0080] Based on the JT-3 and JT-8 peptide sequences of Example 1 and codon usage data, the oligonucleotides oFS5665 (SEQ ID NO:4): 5′-AGYAAYTTYTAYGAYCTSTA-3′ and oFS5667 (SEQ ID NO:5): 5′-CTYTCYTCRTCSAGRTARAA-3′ were constructed on an ABI 392 DNA / RNA Synthesizer and used as primers in a polymerase chain reaction (PCR).
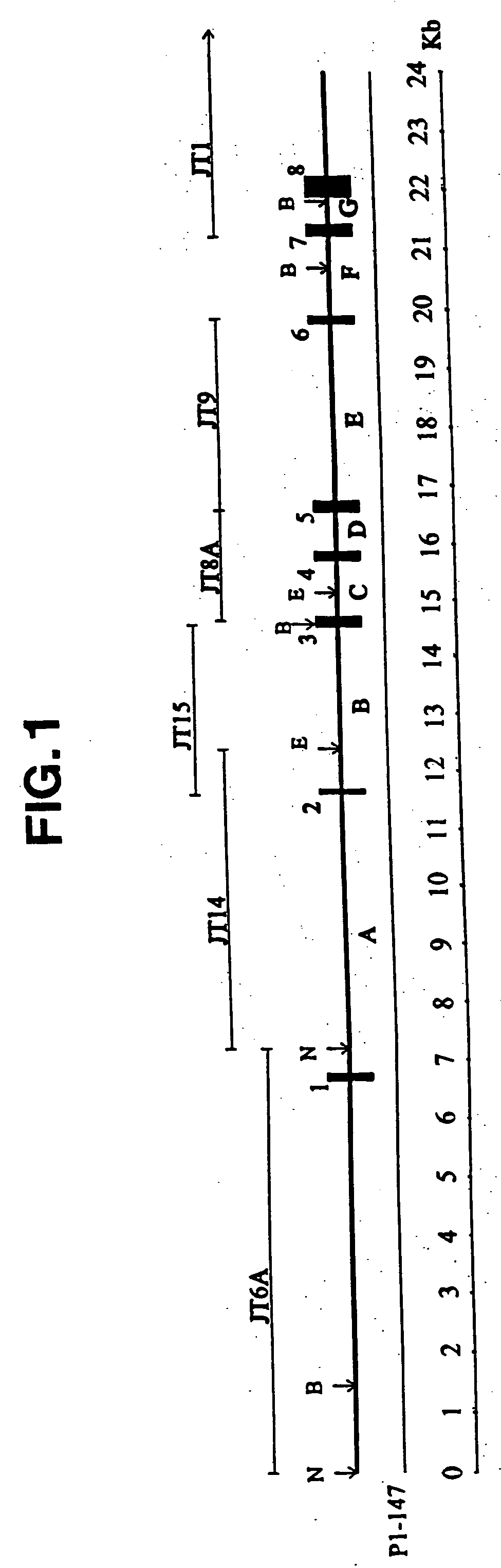
[0081] A human fetal eye Charon BS cDNA library (obtained from Dr. A. Swaroop of the Kellog Eye Institute) was amplified once (Sambrook et al., Molecular Cloning: A Laboratory Manual, 2nd ed., Cold Spring Harbor Press, Cold Spring Harbor, N.Y. (1989)) and screened by PCR (Friedman et al., Screening of λgt11 Libraries, In: PCR Protocols: A Guide to Methods and Applications, Innis et al., eds., Academic Press, NY (1990), pp. 253-260) using a Techne thermal cycler a...
example 3
[0085] This example describes the construction of an expression vector for the production of recombinant PEDF.
[0086] An expression vector was constructed using the plasmid πFS17, which contains the full-length cDNA for human PEDF as described in Example 2. The PEDF coding sequence was placed under the control of a bacteriophage lambda PL promoter present in the plasmid pEV-vrf2 (Crowl et al., Gene, 38, 31-38 (1985)) to obtain the vector pEV-BH. This was accomplished by obtaining a BamH I-Hind III fragment of πFS17 comprising a portion of the PEDF coding region (namely, nucleotide 245 to 1490 of SEQ ID NO:1), digesting plasmid pEV-vrf2 with EcoR I-Hind III, rendering both fragments blunt by means of a fill-in reaction at the BamH I and EcoR I ends with DNA polymerase I (Klenow fragment), and ligating the resultant blunt-ended / compatible-ended fragments to each other. The resultant vector pEV-BH places a distance of 8 nucleotide between the Shine-Dalgarno (SD) sequence and the PEDF c...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com