Single copy genomic hybridization probes and method of generating same
a single-copy, genomic technology, applied in the field of single-copy genomic hybridization probes and method of generating same, can solve the problems of reducing the sensitivity of existing probes, 100 copies of multi-copy repetitive sequences, and essentially benign, serious or even lethal, and achieve the effect of eliminating spurious hybridization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Development of HIRA Gene Probe
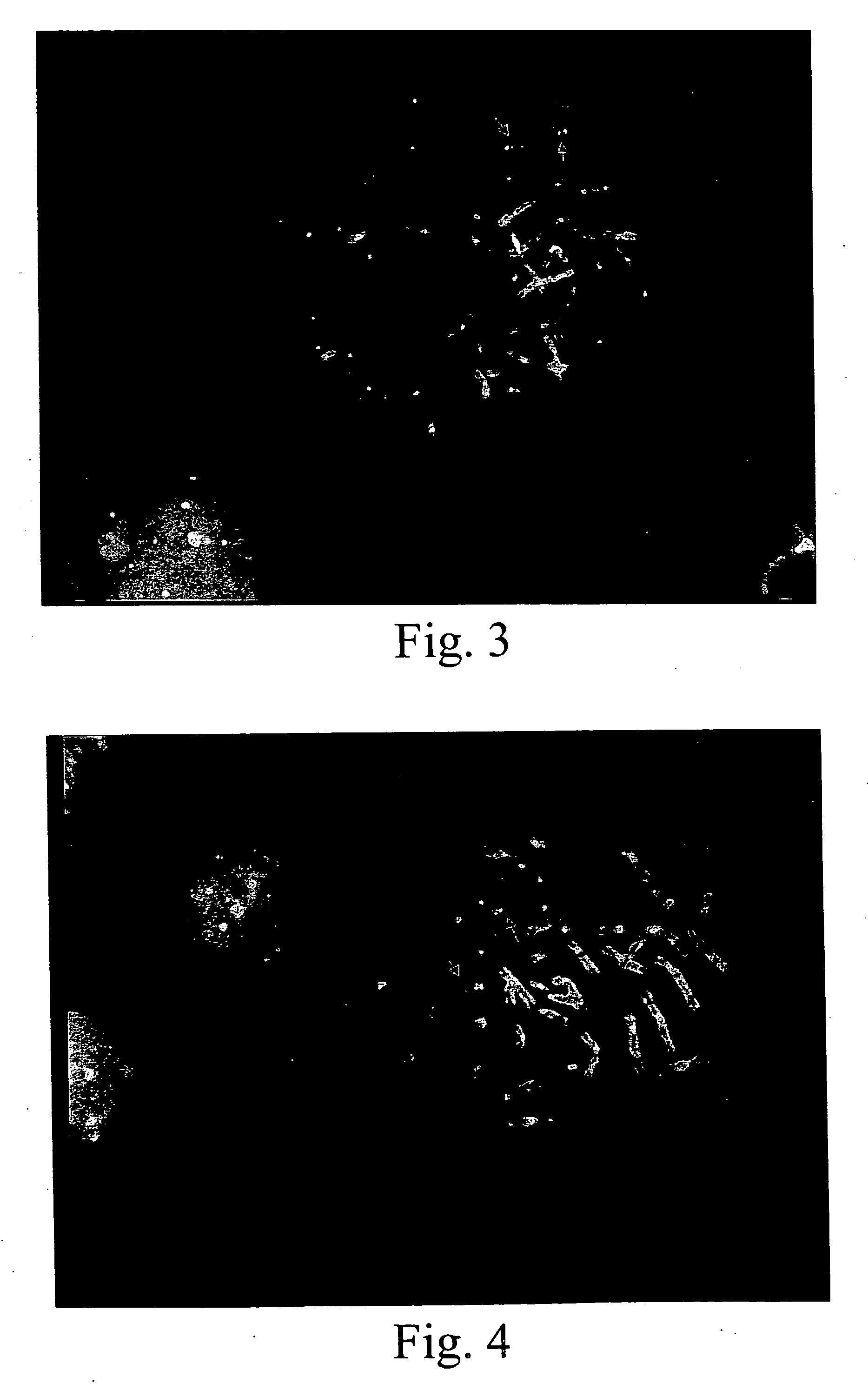
[0068] A known genetic disorder on human chromosome 22 involves a deletion of one HIRA gene in chromosome band 22q11.2, i.e., in normal individuals ; there are two copies of the HIRA gene, whereas in affected individuals, only one copy is present. This deletion is considered to be a cause of haploinsufficiency syndromes such as DiGeorge and Velo-Cardio-Facial Syndromes (VCFS), because insufficient amounts of gene product(s) may disrupt normal embryonic development (Fibison et al., Amer. J. Hum. Genet., 46:888-95 (1990); Consevage et al., Amer. J. Cardiol., 77:1023-1205 (1996)). Other syndromes including Cat Eye Syndrome and derivative chromosome 22 syndrome result from an excess of genomic sequences from this region (Mears et al., Amer. J. Hum. Genet., 55:134-142 (1994); Knoll et al., Amer. J. Med. Genet., 55:221-224 (1995)). Typically individuals with these syndromes have supernumerary derivative chromosome 22s.
[0069] Initially, a computer-based sear...
example 2
Development of NECDIN and CDC2L1 Gene Probes
[0093] The techniques described in Example 1 were used to develop a series of probes for detecting known genetic disorders on chromosome 1 (Monosomy 1p36.3 syndrome; Slavotinek et al.; J. Med. Genet., 36:657-63 (1999)) and on chromosome 15 (Prader-Willi and Angelman Syndromes). Approximately 70% of patients with Prader-Willi or Angelman syndrome exhibit hemizygous deletions of the sequence containing the NECDIN gene (Knoll et al.; Amer. J. Med. Genet., 32:285-290 (1989); Nicholls et al., Amer. J. Med. Genet., 33:66-77 (1989)). The presence of excess copies of this gene is diagnostic for an abnormal phenotype in patients with interstitial duplication or a supernumerary derivative or dicentric chromosome 15 (Cheng et al., Amer. J. Hum. Genet., 55:753-759, 1994; Repetto et al., Am. J. Med. Genet., 79:82-89, 1998). The following Table 3 sets forth the deduced single copy intervals, PCR primer coordinates, SEQ ID Nos. and the lengths of the re...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Tm | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com