Assay for the identification of microorganisms
A microbial and electrochemical technology, applied in the field of analysis for identifying microorganisms, which can solve problems such as unreliability and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Respiratory cycle activity in the presence of effector compounds
[0029] In this example, the assay uses the following steps: (1) The method involves mixing 200 μL of bacterial culture with 1300 μL of buffer (with or without effector and 5 μM DCIP) at a fixed temperature (35° C.) The electrode was mixed in the electrochemical cell for a fixed time (10 min); (2) followed by the addition of mediator (500 μL of 0.2M potassium ferricyanide) at a fixed temperature (35°C) for a fixed time (10 min); and (3) The current is measured at a fixed voltage difference (-100mV) and for a fixed time (120sec). Ferricyanide solution was added such that the final concentration of ferricyanide was 50 mM.
[0030] In this configuration, the analysis uses approximately 0.017cm each 2 surface area of the two platinum electrodes. The measured current is proportional to the concentration of the least concentrated redox pair species in solution. In the present system, there is a large exce...
Embodiment 2
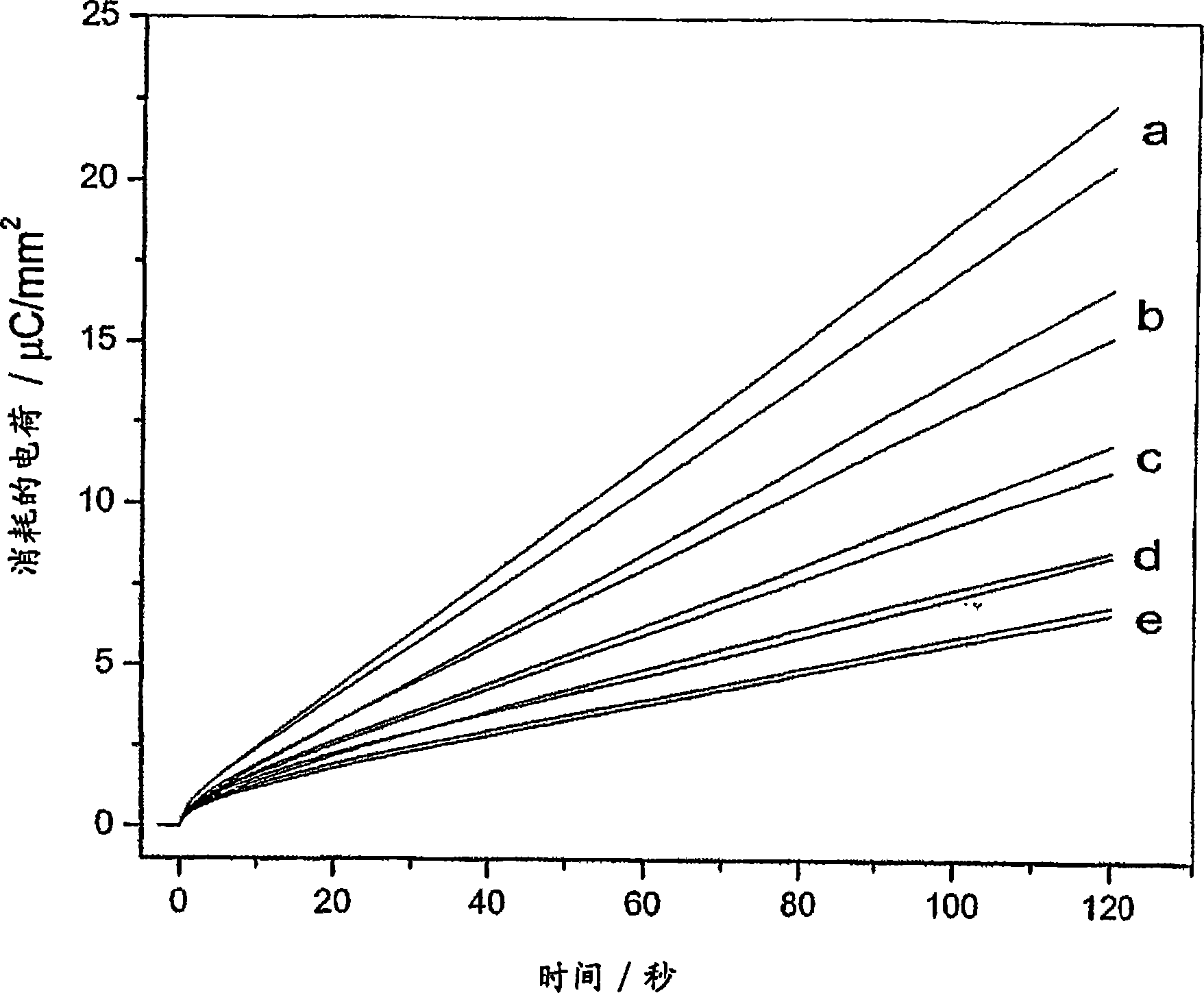
[0042] Distinguish between Gram-positive and Gram-negative bacteria
[0043] The following steps were used for each experiment: (1) Preheat 150 μL aliquots of buffer containing 1 mM glucose (glc) with or without 5 μM DCIP at a fixed temperature (35° C.) for a fixed time (4 min); (2 ) then add 50 μL of bacterial suspension to the sample at a fixed temperature (35°C) for a fixed time (10 min); (3) then add mediator (50 μL 0.4M ferricyanide Potassium) for a fixed time (10 min); and (4) current was measured at a fixed temperature (35° C.) and for a fixed time (120 sec) at a fixed voltage difference (100 mV) between two platinum electrodes. The final ferricyanide concentration in the sample prior to reaction with the microorganism was 40 mM.
[0044] In this configuration, the analysis uses approximately 0.03cm each 2 surface area of the two platinum electrodes. The measured current depends on the concentration of the least concentrated redox species in solution. In our syste...
Embodiment 3
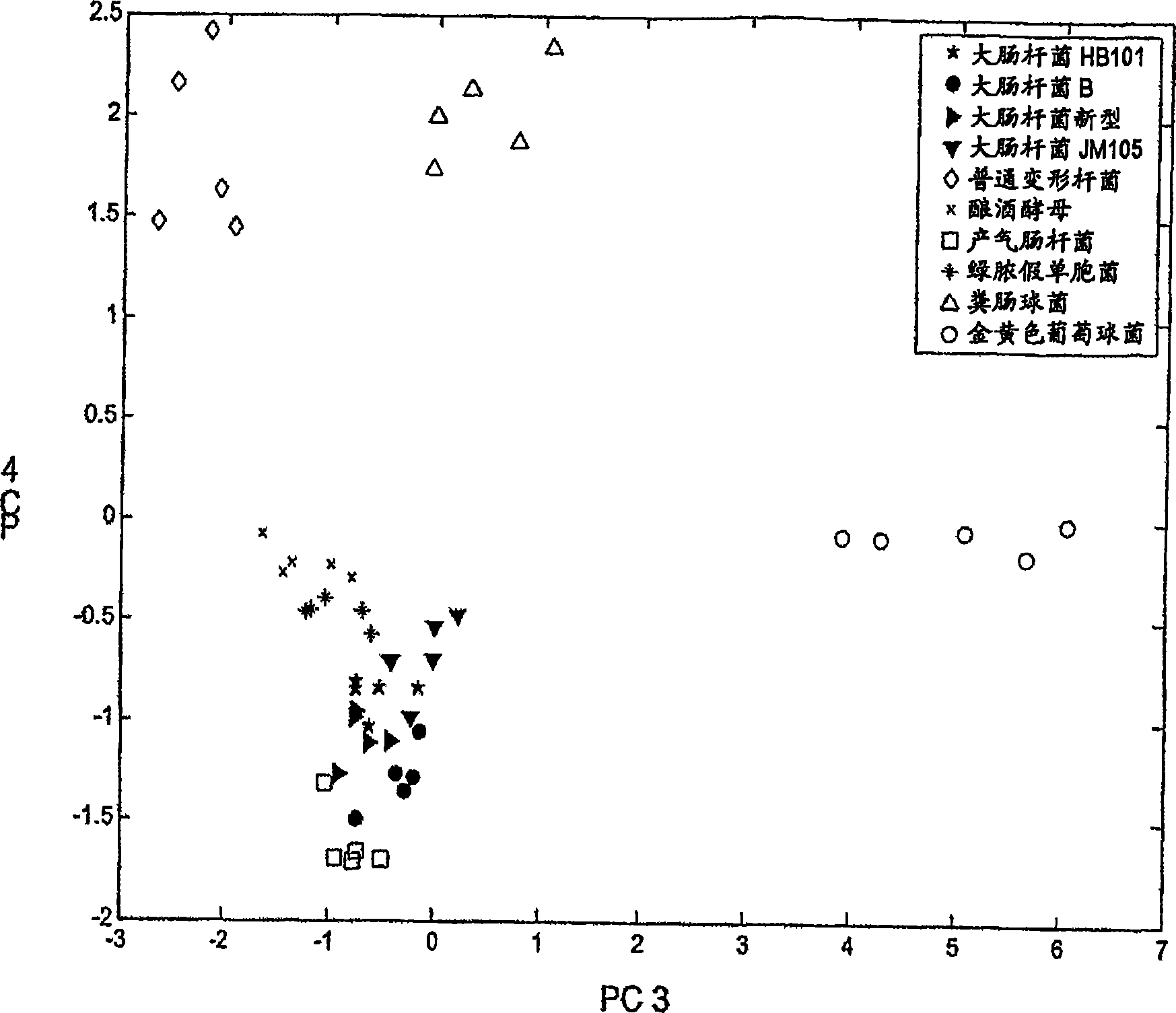
[0051] Discrimination of 10 microbial strains using pattern recognition by principal component analysis of respiratory activity data
[0052] In this example, individual microorganisms were subjected to 22 different effector compounds in addition to control measurements. The following steps were used for each experiment: (1) Preheat 150 μL aliquots of buffer containing the effector compound and 5 μM DCIP at a fixed temperature (35° C.) for a fixed time (4 min); (2) subsequently at a fixed temperature ( Add 50 μL of bacterial suspension to the sample at 35°C) for a fixed time (10 min); (3) then add mediator (50 μL 0.4M potassium ferricyanide) at a fixed temperature (35°C) for a fixed time (10 min); and (4) current was measured at a fixed temperature (35° C.) and for a fixed time (120 sec) at a fixed voltage difference (100 mV) between two platinum electrodes. The final ferricyanide concentration in the sample prior to reaction with the microorganism was 40 mM. For each microo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com