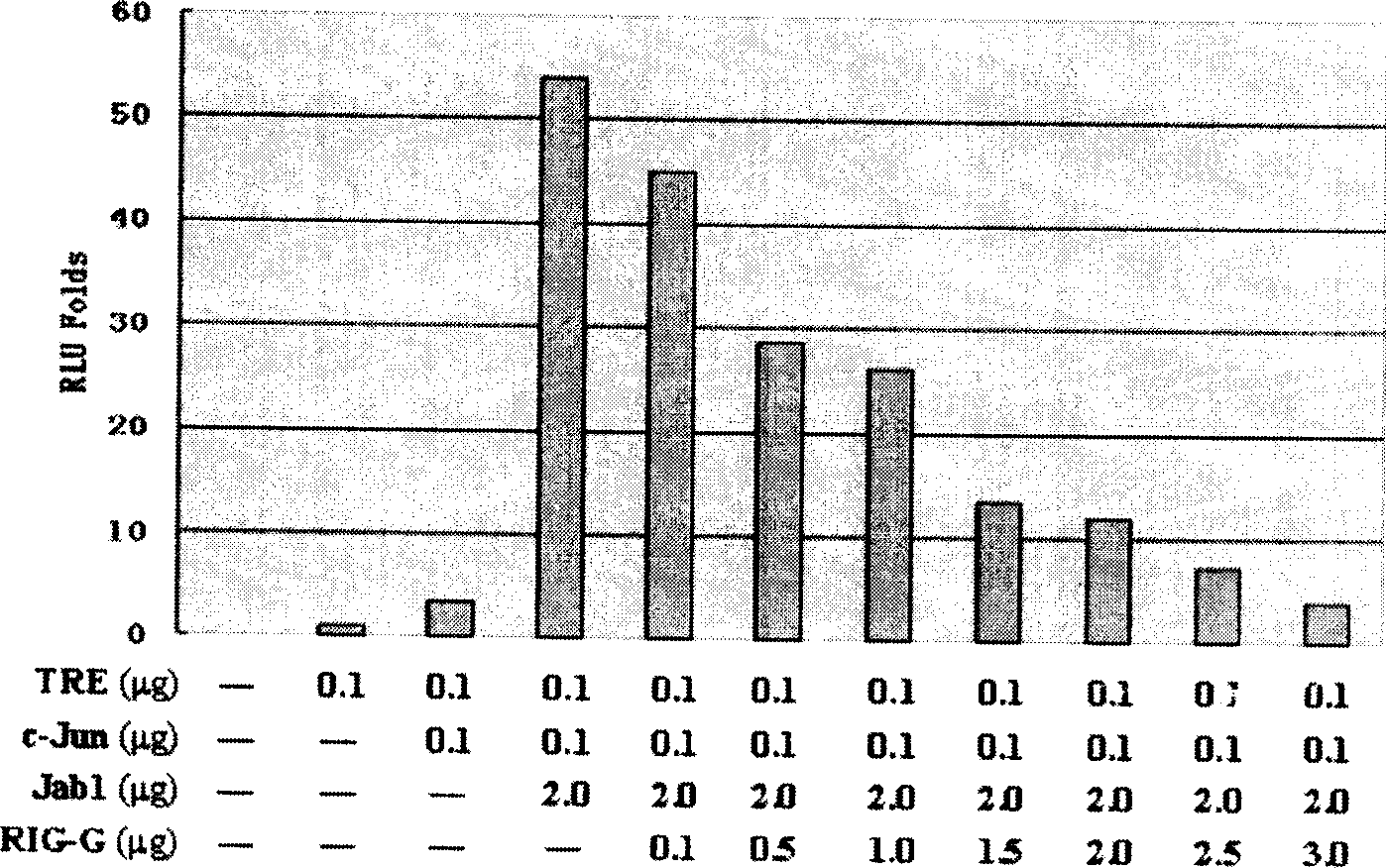
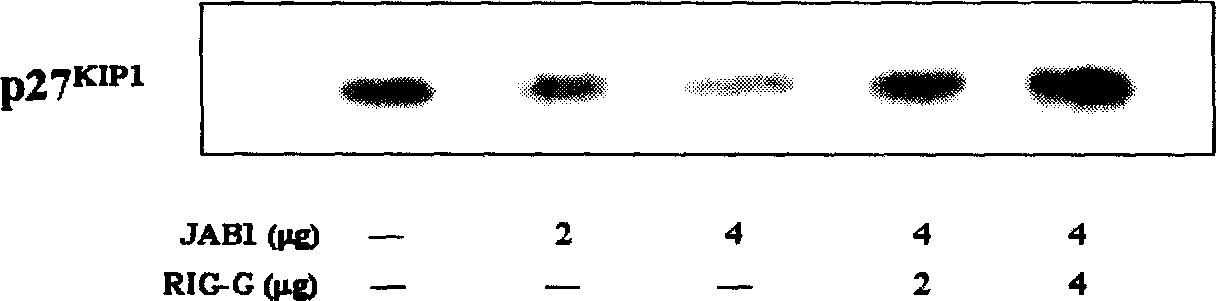
High efficiency siRNA molecule to target gene Rig-G
A gene and purpose technology, applied in the field of small double-stranded RNA molecules, can solve the problems of inhibiting AP-1 protein, affecting binding, and reducing stability.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] High-efficiency siRNA to inhibit expression of target gene Rig-G (1)
[0031] 1. Select the sequence on the target gene and synthesize the DNA template:
[0032] According to the basic characteristics of effective RNAi sequences, we selected sites in the cDNA sequence of the target gene Rig-G gene (see Table 1 and Table 2) and synthesized DNA templates (synthesized by Boya Biotech).
[0033] Basic requirements for selecting candidate siRNA sequences on target genes:
[0034] 1) Search for a candidate sequence (but not absolute) at a position 50-100 bp downstream of the promoter of the coding region of the target gene.
[0035] 2) The two template strands (forward strand and anti-strand) of the siRNA duplex: both start with AA to ensure that the 3' of the siRNA duplex transcribed in vitro protrudes two UUs.
[0036] 3) Among the paired 19 nucleotides, the first position is G, and the nineteenth position is C.
[0037] 4) The GC content is preferably 40-55%.
[0038] ...
Embodiment 2
[0061] High-efficiency siRNA to inhibit the expression of target gene Rig-G (2)
[0062] 1. Select the sequence on the target gene and synthesize the DNA template:
[0063] According to the basic characteristics of effective RNAi sequences, we selected sites in the cDNA sequence of the target gene Rig-G gene (see Table 1 and Table 2) and synthesized DNA templates (synthesized by Boya Biotech).
[0064] Basic requirements for selecting candidate siRNA sequences on target genes:
[0065] 1) Search for a candidate sequence (but not absolute) at a position 50-100 bp downstream of the promoter of the coding region of the target gene.
[0066] 2) The two template strands (forward strand and anti-strand) of the siRNA duplex: both start with AA to ensure that the 3' of the siRNA duplex transcribed in vitro protrudes two UUs.
[0067] 3) Among the paired 19 nucleotides, the first position is G, and the nineteenth position is C.
[0068] 4) The GC content is preferably 40-55%.
[00...
Embodiment 3
[0090] Efficient siRNA to inhibit expression of target gene Rig-G (3)
[0091] 1. Select the sequence on the target gene and synthesize the DNA template:
[0092] According to the basic characteristics of effective RNAi sequences, we selected sites in the cDNA sequence of the target gene Rig-G gene (see Table 1 and Table 2) and synthesized DNA templates (synthesized by Boya Biotech).
[0093] Basic requirements for selecting candidate siRNA sequences on target genes:
[0094] 1) Search for a candidate sequence (but not absolute) at a position 50-100 bp downstream of the promoter of the coding region of the target gene.
[0095] 2) The two template strands (forward strand and anti-strand) of the siRNA duplex: both start with AA to ensure that the 3' of the siRNA duplex transcribed in vitro protrudes two UUs.
[0096] 3) Among the paired 19 nucleotides, the first position is G, and the nineteenth position is C.
[0097] 4) The GC content is preferably 40-55%.
[0098] 5) Homol...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com